Abstract
Self-incompatibility (SI) is a widespread mechanism in flowering plants which prevents self-fertilization and inbreeding. In Brassica, recognition of the highly polymorphic S-locus cysteine-rich protein (SCR; or S-locus protein 11) by the similarly polymorphic S-locus receptor kinase (SRK) dictates the SI specificity. Here, we report the crystal structure of the extracellular domain of SRK9 (eSRK9) in complex with SCR9 from Brassica rapa. SCR9 binding induces eSRK9 homodimerization, forming a 2:2 eSRK:SCR heterotetramer with a shape like the letter “A”. Specific recognition of SCR9 is mediated through three hyper-variable (hv) regions of eSRK9. Each SCR9 simultaneously interacts with hvI and one-half of hvII from one eSRK9 monomer and the other half of hvII from the second eSRK9 monomer, playing a major role in mediating SRK9 homodimerization without involving interaction between the two SCR9 molecules. Single mutations of residues critical for the eSRK9-SCR9 interaction disrupt their binding in vitro. Our study rationalizes a body of data on specific recognition of SCR by SRK and provides a structural template for understanding the co-evolution between SRK and SCR.
Similar content being viewed by others
Introduction
In flowering plants, self-incompatibility (SI) is a universal mechanism for avoidance of self-fertilization and inbreeding, thus maintaining their genetic diversity1. In the Brassicaceae family, SI is mediated by variant haplotypes of a single highly polymorphic genetic locus, termed the S locus2, which generally contains three highly polymorphic genes, the stigma-expressed S-locus receptor kinase (SRK), the pollen-expressed S-locus cysteine-rich protein (SCR; or S-locus protein 11) and the S-locus glycoprotein (SLG)3,4,5,6,7. Genetic and biochemical studies established SRK and SCR as the sole determinants of SI specificity, and SRK as the receptor for SCR, which allows the stigma to discriminate between “self” and “non-self” pollen in the SI response4,8,9,10,11. Because an SCR protein will only bind and activate the SRK variant encoded in the same S-locus haplotype, the SRK and SCR proteins must co-evolve to maintain their interaction. This highly specific recognition of “self” SCR by the extracellular domain of SRK (eSRK) induces activation of the SRK kinase, consequently triggering a signaling cascade for inhibition of the “self” pollen9,12. SCR-enhanced SRK homodimerization is important for initiation of this signaling12,13,14.
SRK belongs to the large family of receptors in plants, designated receptor kinases (RKs), which consist of more than ten subfamilies that play important roles in diverse biological processes15. SRK is the prototypic member of the S-domain RLK (SD-RLK) subfamily with ∼40 encoded in the Arabidopsis thaliana genome16. The eSRK is characterized by two contiguous N-terminal lectin-like domains followed by a region containing 12 conserved cysteine residues17. Structural modeling predicted the six N-terminal cysteines to be contained within an EGF-like domain and the six C-terminal cysteines within a PAN/APPLE domain17. Comparison of eSRK sequences identified several hyper-variable (hv) regions, designated hvI, hvII and hvIII, which were predicted to be important for SI specificity18,19. Consistently, studies of a small number of variants demonstrated that residues from the hvI and hvII regions are required for “self” SCR recognition20,21. But whether these regions are generally required for the recognition of all SCRs by their cognate SRKs remains unknown. In the case of SCR variants, which are more polymorphic than SRKs, an NMR study suggested that these small proteins of ∼50 amino acids that typically contain eight conserved cysteines may all have a structure similar to defensins22. One study of two B. oleracea SCR variants, SCR6 and SCR13, showed that four contiguous amino-acid residues located between the fifth and sixth conserved cysteines are critical for the specific recognition of SCR13 by SRK13 but not for the recognition of SCR6 by SRK6 in planta23. Thus, no general rules have yet emerged for predicting which residues in receptor and ligand determine their specific interaction.
To bridge the gap caused by the lack of structural mechanism underlying SRK recognition of SCR, we solved the crystal structure of eSRK9-SCR9 complex, which turns out to be a 2:2 eSRK:SCR heterotetramer having a shape like the letter “A”. All three hv regions of eSRK9 mediate the specific recognition of SCR9. Interestingly, SCR9 induces the dimerization of eSRK9 by interacting with one half of hvII from one eSRK9 monomer and the other half of hvII from the second eSRK9 monomer, without involving interaction between the two SCR9 molecules. Together, these findings for the first time elucidate the molecular mechanism of SI in the Brassicaceae family.
Results
Overall structure of the eSRK9-SCR9 heterotetrameric complex
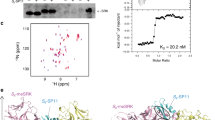
Several pairs of eSRK and SCR proteins from Brassica rapa were expressed in insect cells and screened for protein purification and crystallization. In the end, eSRK9 and SCR9 were found to be well expressed and to form a stable complex as indicated by a gel filtration assay. Indeed, this assay showed that, in the presence of SCR9, the eSRK9-containing fraction was shifted from a molecular weight of ∼50 kD to a molecular weight of ∼120 kD, which approximates the weight of a dimeric complex of the two proteins, indicating that SCR9 induced eSRK9 dimerization in solution (Figure 1A). The crystal structure of the complex was solved with single wavelength anomalous dispersion (SAD) using an iodine derivative crystal (Supplementary information, Table S1).
SCR9 induces eSRK9 homodimerization. (A) SCR9 induces eSRK9 homodimerization in vitro. Left, superposition of the gel filtration chromatograms of the eSRK9, SCR9, and eSRK9+SCR9 samples. The vertical and horizontal axes represent UV absorbance (280 nm) and elution volume (ml), respectively. Right, coomassie blue staining of the peak fractions shown on the left following SDS–PAGE. (B) eSRK9 and SCR9 form a heterotetramer in crystal. Shown are the overall crsytal structures of the eSRK9-SCR9 complex in two different orientations. “N” and “C” represent the N- and C-termini, respectively. Structural domains of eSRK9 are labeled. Schematic representation of structural domains of SRK9 is shown above the structure. The positions of 12 cysteins are indicated. hv, hyper-variable region; SP, signal peptide; TM, transmembrane.
The eSRK9-SCR9 complex contains an eSRK9 dimer enveloping two SCR9 molecules, yielding a 2:2 tetrameric complex (Figure 1B). The overall structure of the complex resembles the letter “A” with the two eSRK9 monomers corresponding to the two stems and the two SCR9 molecules to the bar of the letter. The EGF-like domains and the second lectin domains of the two eSRK9 monomers form a large hydrophobic pocket for SCR9 recognition (Figure 1B). These domains appear to play central roles in the formation of the tetrameric complex, as they are involved in both SCR9 recognition and eSRK9 homodimerization, which is both ligand- and receptor-mediated. The two eSRK9 monomers contribute to recognition of SCR9 by sandwiching the ligand through the second lectin domain of each the two eSRK9 monomers. The EGF-like domain from one eSRK9 monomer further strengthens the eSRK9-SCR9 interaction by contacting another side of SCR9. The homodimerization of eSRK9 is mediated by the interactions between the two lectin domains and the contacts between the looped-out helix of the EGF-like domain from one eSRK9 monomer and the second lectin domain from the other monomer (Figure 1B). The two SCR9 molecules are positioned far away from each other and no contacts are made between them.
The S-domain architecture of eSRK9
SRK9 displays an elongated structure comprising several clearly defined domains (Figure 2A). Both of the two N-terminal lectin domains are globular, each having the structure of a nine-stranded β-barrel (Figure 2B). The configuration of the β-strands in both of these domains has an approximate three-fold internal symmetry, forming a “Y” or trefoil-shaped structure. As previously predicted17, a database search using the DALI server revealed that these two lectin domains share appreciable structural similarity to a mannose-recognizing lectin from Japanese Marasmius oreades24 despite their weak sequence similarity to the latter lectin (Figure 2B and Supplementary information, Figure S1A). However, the hairpin loops of these proteins vary greatly in length, sequence, and conformation (Figure 2B and Supplementary information, Figure S1A). In the nine-stranded barrels, each layer contains inward-pointing hydrophobic residues that are likely important for formation of the fold. Interestingly, structure-based sequence alignment showed that these residues are highly conserved in the first lectin but not in the second lectin domain among the S-domain containing RKs of A. thaliana (Supplementary information, Figure S2). Located at outer surface of the second lectin domain, Trp178 and Trp280 of eSRK9, which are important for the interaction between the two lectin domains (Supplementary information, Figure S1B), are also conserved (Supplementary information, Figure S2), suggesting that these S-domain proteins may share a similar structure.
Structure of eSRK9. (A) Overall structure of eSRK9 shown in cartoon. (B) Structural alignment of the two N-terminal lectin domains of eSRK9 with Narcissus pseudonarcissus lectin (NPL; PDB code: 4TKC). (C) Structural alignment of the EGF-like domain of eSRK9 with EGF (PDB code: 1EPG). (D) Structural alignment of the HGF-like domain of eSRK9 with NK1 (PDB code: 1GMO).
A modeling study had suggested that residues 293-346 of eSRK6 encode an EGF-like domain17 that is principally defined by six cysteines, which are known to form disulphide bonds in a 1-3, 2-4, and 5-6 pattern25. Indeed, this region of eSRK9 forms a structure that can be largely aligned with that of EGF26 (Figure 2C). In addition, a short α-helix is embedded within the domain, and Trp326 from this helix tightly packs against the Cys298-Cys312 and Cys314-Cys335 disulfide bonds (Figure 2C). Following the EGF-like domain, the predicted PAN/APPLE domain17 is mainly composed of a 5-stranded anti-parallel β-sheet with an α-helix packing at one side and two long loops at the other. Supporting the sequence-based prediction, structural comparison revealed that this structural domain is most similar to the N-terminal domain of hepatocyte growth factor (HGF) (Figure 2D), which is a member of the PAN/APPLE family protein and responsible for heparin binding27. We therefore designate this domain as an HGF-like domain. Despite the similar fold, eSRK9 lacks the critical residues important for HGF binding to heparin. In eSRK9, the two long loops from the HGF-like domain engage in interactions with both the second lectin domain and the EGF-like domain, which together with inter-domain interactions between the two lectin domains result in an elongated eSRK9 structure (Figure 2A).
The structure of SCR9
SCR9 is a small cysteine-rich peptide and adopts a compact globular fold with a long α-helix tightly packing against the 3-stranded anti-parallel β-sheet at one side (Figure 3A and 3B). The three disulfide bonds, Cys40-Cys65, Cys48-Cys74, and Cys63-Cys76, appear critical for the stabilization of the α/β motif (Figure 3A and 3B). Tyr52, which is conserved in many Brassica SCRs (Supplementary information, Figure S3A), is sandwiched between the α-helix and the β-sheet (Supplementary information, Figure S3B), and this configuration may also contribute to the structural integrity of SCR9. Database searches revealed that the structure of SCR9 is similar to the structures of SCR822, plant defensins28, and scorpion neurotoxins29 (Figure 3C), though the positions of their disulfides bonds are not conserved. Despite this high level of structural similarity, these proteins exhibit strikingly different surfaces (Supplementary information, Figure S3C), which may allow them to have different mechanisms of action and biological activities.
Structure of SCR9. (A) Primary sequence of SCR9. Fragment that could be visuallized in the structure is colored by purple, the missing parts are colored by gray. The three disulfide bonds are marked by blue lines, the fourth theotically existing disulfide bond is marked by dashed orange line. (B) Overall structure of SCR9 shown in cartoon. Three disulfide bonds are shown in yellow. (C) Structural alignment of SCR9, SCR8 (PDB code: 1UGL), Pisum sativum defensin 1 (PSD1) (PDB code: 1JKZ), and BmK M1 (PDB code: 1SN1).
The mechanism underlying specific recognition of SCR9 by eSRK9
In the eSRK9-SCR9 complex, the interaction of one SCR9 molecule with the eSRK9 dimer buries 2 547 Å2 of solvent-accessible surface. About half (1 190 Å2) of the buried surface comes from contacts of the short hairpin loop of SCR9 with the β-barrel of one eSRK9 monomer and the loop linking the second lectin domain to the EGF-like domain of the same eSRK9 molecule (Figure 4A). At the center of this interface is Phe69 of SCR9, which interacts with a hydrophobic cavity formed by Val211, Phe267, Pro287, and Phe290 of SRK9 (Figure 4B). Additional hydrophobic interactions of this interface are established between Tyr73 of SCR9 and its neighboring residues of SRK9. Several hydrogen bonds involving Thr66, Tyr73, and Asp75 of SCR9 further fortify interactions of this interface. The short α-helix of the second lectin domain from the other eSRK9 monomer interacts with the long α-helix of SCR9 mainly through hydrophobic contacts (Figure 4C). Both Leu277 and Val278 of SRK9 engage hydrophobic interactions with Phe51 of SCR9. The carbonyl oxygen atoms of these two SRK9 residues also make a bidentate hydrogen bond with Arg72 of SCR9, which in turn packs against the side chain of Ile279 of SRK9. Distal to the interface is Pro282 of SRK9 that tightly packs against Gly38 of SCR9. The looped-out region of the EGF-like domain and the linker between the EGF-like domain and second lectin domain interact with the C-terminal end of the α-helix and one β-strand of SCR9, respectively (Figure 4D). The former contact is mainly mediated by polar interactions. In addition to forming a pair of salt bridges with Glu325 and Asp330 of SRK9, Lys55 of SCR9 also establishes hydrophobic contacts with Thr332 and Arg333 of SRK9 through its aliphatic portion. Arg333 of SRK9 further enhances interactions of this region by hydrogen bonding the carbonyl oxygen atoms of Asn54 and Lys55 of SCR9. Structure-based sequence alignment revealed that residues from the hv regions of eSRK9 govern its interaction with SCR9, and that hvII forms the densest interactions with SCR9 (Figure 4E and Supplementary information, Figure S4).
Recognition mechanism of SCR9 by eSRK9. (A) Hyper-variable regions (highlighted in red) mediate eSRK9 interaction with SCR9. Details of the regions highlighted in blue, black and pink frames are shown in B, C and D, respectively. (E) Structural based sequence alignment of eSRKs from Brassica rapa around the hyper-variable regions of SRK9. Above the alignment, purple dots mark the residues that are involved in interaction with SCR9 and cyan dots mark the residues involved in eSRK9 homodimerization.
Structural basis for SCR9-induced eSRK9 dimerization
SCR9 contributes to eSRK9 dimerization via its simultaneous interaction with the second lectin domains of the two eSRK9 monomers (Figure 1B). In addition, eSRK9 dimerization is also mediated by direct interaction between the two eSRK9 monomers. A hairpin loop of the EGF-like domain from one monomer interacts with a loop of the HGF-like domain from the other monomer, forming one eSRK9 homodimerization interaction interface (Supplementary information, Figure S5A). Interaction of this interface is mediated by Van der Waals contacts made by Asn307 from the EGF-like domain with Ala412 and Val413 from the HGF-like domain. In addition, the loops C-terminal to the last β-strand from the second lectin domains of the two eSRK9 monomers also contribute to homodimerization. The Phe290 residues from the two eSRK9 monomers stack against each other, whereas Gln291 from one monomer establishes a hydrogen bond with the carbonyl oxygen of Ala288 from the other monomer (Supplementary information, Figure S5A). Another homodimerization interface comprises the looped-out helix of the EGF-like domain from one eSRK9 monomer and an anti-parallel β-sheet of the second lectin domain from the other monomer (Supplementary information, Figure S5B), which form a combination of polar and hydrophobic interactions.
Mutations disrupting the interaction between SCR9 and eSRK9
Our crystal structure is supported by our observation that SCR9 induced eSRK9 homodimerization in solution (Figure 1A). To further verify our structural observations, we mutated residues located at the interfaces between SCR9 and eSRK9 and tested the impact of these mutations on SCR9-induced eSRK9 homodimerization using gel filtration. Substitutions of several residues located at the interface between the short hairpin loop of SCR9 and the β-barrel of eSRK9 (Figure 4B) were found to be disruptive. Thus, mutating the F69 residue in SCR9 to the charged residue Glu resulted in loss of the ability of SCR9 to induce eSRK9 homodimerization. Similarly, mutations of V211 and P294 of SRK9 to the equivalent residues in SRK8 (E and M, respectively; Figure 4E) abolished SCR9-induced eSRK9 homodimerization (Figure 5A), consistent with the expectation that the V211E and P294M mutations would generate steric effects due to the limited space around these two residues. Interestingly, and supporting the significance of the P294 residue of SRK9 in the SI response, a mutation of its equivalent residue, V301, in Arabidopsis lyrata SRK25 was previously shown to greatly weaken the ability of this variant to inhibit SCR25 pollen in vivo21. Further confirmation of our crystal structure was obtained by substituting H331, T332, and R333 of eSRK9 with their equivalent residues (R331, S332, and S333 in SLG9, respectively; Supplementary information, Figure S6) in SLG9, a protein that shares 98% sequence identity with eSRK9 but does not interact with SCR913. These mutations abolished the eSRK9-SCR9 interaction (Figure 5A), as predicted from the structure of the eSRK9-SCR9 complex. Indeed, the T332R and R333S mutations would directly compromise the interaction of eSRK9 with SCR9 (Figure 5B), while substitution of H331 with the bulkier Arg residue may perturb the local conformation of eSRK9 (Figure 5B), thus further reducing its interaction with SCR9.
Mutagenesis analysis of the eSRK9-SCR9 complex. (A) Superposition of the gel filtration chromatograms of the wild-type eSRK9 and SCR9 with their respective mutants. Right, coomassie blue staining of the peak fractions shown on the left following SDS–PAGE. The assays were performed as described in Figure 1A. (B) H331, T332 and R333 of SRK9 mediate inter- and intra-molecular interactions.
Discussion
The data presented here demonstrate that eSRK9 is sufficient for recognition of SCR9, forming a 2:2 eSRK9-SCR9 complex, and further support previous observations. The crystal structure of this complex represents the first structure of ligand-induced homodimer of an RK. In addition to SCR9, homodimerization of eSRK9 may be further strengthened by its trans-membrane segment, which could be responsible for the observed pre-formed SRK homodimers13,30. The mode of SCR9-induced eSRK9 homodimerization differs from those of ligand-induced homodimerization of receptor tyrosine kinases (RTKs) in mammals. In the latter cases, homodimerization results from either cross-linking of an RTK by a dimeric ligand or ligand-induced conformational changes in the RTK31. By comparison with RTKs, SRK9 homodimerization, though also contributed by the direct contacts between two SRK9 molecules, is mainly mediated by two independent SCR9 molecules. Despite this difference, our data support the idea that ligand-induced dimerization is important for the activation of an RK32,33. The dimerization model suggests that interaction between an SRK and an SCR may not always be translated into activation of the SRK, because their interaction may not necessarily result in receptor homodimerization. This possibility is consistent with the observations that non-cognate SRK-SCR binding could be detected20 and that an SCR6 mutant interacted with SRK6 but failed to induce an SI response in planta23. This is also structurally possible, because SCR9 recognition by a monomeric eSRK9 is mediated mainly through hvI, the C-terminal half of hvII, and hvIII, and is strengthened by interaction with the N-terminal half of hvII from the other SRK9 monomer. Thus, SRK9 homodimerization may enhance signaling specificity in the SI response.
Our structure rationalizes previous biochemical and functional data related to specificity in the SRK-SCR interaction. While the eSRK9-interacting residues from both the α-helix and β-sheet of SCR9 are relatively scattered along the length of this protein (Supplementary information, Figure S3A), nearly all of the SCR9-interacting residues in eSRK9 are located in the three hv regions (Figure 4E and Supplementary information, Figure S4), supporting an important role of these regions in SCR recognition as previously suggested2,12,18,19,20,21. Although all three hv regions are involved in SCR9 recognition, the hvI and hvII regions contribute the majority of SRK9 interaction with SCR9. This structural observation agrees well with a previous in vivo study of two SRK variants, CgSRK7 and AlSRK25, which demonstrated that residues from these two regions were necessary for activation of the SI response21. Among the three SRK9-SCR9 interfaces, the densest interactions come from contacts of the residues between the fifth and sixth cysteines of SCR9 with hvI and hvII, which explains the importance of this region for recognition specificity in other SCR variants34. Collectively, these results suggest that SRK recognition of SCR shares a conserved mechanism and the structure of the eSRK9-SCR9 complex solved in this study is applicable to other SRK-SCR pairs. Consistently, modeling studies of two pairs of SRK-SCR complexes, eSRKa-SCRa and eSRK25-SCR25 from A. lyrata, showed that they both formed stable 2:2 tetrameric complexes with a similar recognition mechanism to that of the eSRK9-SCR9 complex (Figure 1B and Supplementary information, Figure S7A). However, in view of the extensive polymorphisms exhibited by SRK and SCR variants, it remains formally possible that the contributions of the three eSRK9-SCR9 interfaces might vary in different SRK-SCR pairs. This possibility is suggested by the fact that the location of SCR residues critical for SRK activation can differ between SCR variants23. Interestingly, structural superposition of the crystal and modeled structures showed that steric clashes exist between an SRK and a non-self SCR but vary in the different interfaces (Supplementary information, Figure S7B), which can be used by SRKs to repel non-self SCRs. Nonetheless, structural mapping of the specificity-determining residues in eSRK9 and SCR9 represents an important step toward understanding the co-evolution between SRK and SCR.
Materials and Methods
Protein expression and purification
The sequences of the Brassica rapa SRK9 extracellular domain (residues 30-436, eSRK9) and SCR9 (residues 25-83) were codon optimized for expression in Trichoplusia ni and synthesized by Genewiz. Constructs of eSRK9 with a C-terminal 6×His tag and SCR9 with a cleavable N-terminal 6×His-SUMO tag were generated by a standard PCR-based cloning strategy and their identities were confirmed by sequencing. Baculoviruses of eSRK9 and SCR9 were constructed with the Bac-to-Bac system (Invitrogen) according to the manufacturer's protocols. All constructs were expressed in High Five insect cells at 22 °C using the pFastBac-1 vector (Invitrogen) with a modified N-terminal hemolin signal peptide. One liter of cells (2.0 × 106 cells /ml) was infected with 20 ml baculovirus and the media was harvested after 48 h. Proteins were purified individually using Ni-NTA (Novagen) and eluted with buffer containing 25 mM Tris, pH 8.0, 150 mM NaCl, and 250 mM imidazole. The Sumo tag was removed from SCR9 before elution by incubating the bound proteins with PreScission protease (GE Healthcare) in a 1:100 molar ratio at 4 °C for 4 h. The eluted proteins were then concentrated and further purified by size-exclusion chromatography (Hiload 16/600 Superdex 200 prep grade, GE Healthcare) in buffer containing 10 mM Bis-Tris, pH 6.0 and 100 mM NaCl. Peak fractions of each protein were pooled together and concentrated to ∼10 mg/ml and flash frozen in liquid nitrogen. To obtain the eSRK9-SCR9 complex, the purified eSRK9 and SCR9 proteins were mixed and incubated at 4 °C for 30 min. The mixture was subsequently subjected to gel filtration (Hiload 16/600 Superdex 200 prep grade, GE Healthcare) in buffer containing 10 mM Bis-Tris, pH 6.0 and 100 mM NaCl. Fractions corresponding to the complex were pooled together and concentrated to ∼10 mg/ml for crystallization.
Crystallization, data collection, structure determination, and refinement
Crystals of the eSRK9-SCR9 complex were generated by the hanging-drop vapor-diffusion method using the commercially available screening kits from Hampton Research. The drops were set up with 1 μl protein plus 1 μl reservoir solution at 18 °C. Initial crystals with poor X-ray diffraction were obtained in buffer containing 0.2 M sodium malonate, pH 7.0, and 20% PEG 3350 within 1 week. Diffraction quality crystals emerged in buffer containing 0.15 M sodium malonate, pH 7.0, 16% PEG 3350, and 4% PEG 400 within 2 weeks. For data collection, the crystals were equilibrated in a cryoprotectant buffer containing reservoir buffer plus 12% (v/v) glycerol. All the diffraction data sets were collected at the Shanghai Synchrotron Radiation Facility (SSRF) on beam line BL17U1 using a CCD detector. The data were processed using HKL2000 software35. Initial phases were obtained by SAD using an iodine derivative of the native crystal, which was obtained by soaking a native crystal in the cryoprotectant buffer supplemented with 0.6 M KI for 100 s before being put on beam. Locations of the iodine atoms and phase calculation were performed using the program SHELXD36. The phases were further improved using the program DM (density modification) included in CCP4. COOT37 and PHENIX38 were used for model building and structure refinement, respectively. All the structure figures were prepared using PyMol (PyMOL).
Gel filtration assay
Gel filtration was performed using a Hiload 16/600 Superdex 200 prep grade (GE Healthcare) pre-equilibrated in buffer containing 10 mM Bis-Tris, pH 6.0, and 100 mM NaCl. The eSRK9 and SCR9 proteins (wild-type or various mutants) were purified as described above. After incubation at 4 °C for 30 min, the eSRK9-SCR9 mixtures were subjected to gel-filtration analysis. The assays were performed with a flow rate of 1 ml/min and an injection volume of 1.5 ml at 4 °C. Samples from relevant fractions were applied to SDS-PAGE and visualized by coomassie blue staining.
Computational studies
The receptor proteins eSRK25 and eSRKa were modeled using JACKAL program39 based on the X-ray solved eSRK9-SCR9 homolog structure with > 60% sequence similarities. The ligand proteins SCR25 and SCRa were modeled using ITASSER program40 based on the program's structure database and the X-ray solved eSRK9-SCR9 structure. The models with the highest scores were picked out as the results.
The complex formed by the interaction between SRK7 and SCR7 was predicted by the docking method HoDock41 which incorporates an initial rigid docking and a refined semi-flexible docking. In this work, the experimental solved tetramer structure eSRK9-SCR9 showing a promisingly similar binding mode, which were both used as restraints for conformational searching and model selection. Totally 8 500 complex structures were generated and scored to pick up the final correct complex structure model.
Molecular dynamics simulation package Gromacs 4.542 with OPLS force field was used for the minimization to relax and equilibrate the structures in solution. Then the minimized structures, without atom clashes, fitting best with stereo-chemical restraints were selected as the built model.
Author Contributions
JC, RM, Zhifu Han and Zehan Hu designed the project. RM performed the experiments of protein purification, crystallization, gel-filtration assays. RM, GL, HZ collected x-ray data. The structure was determined by JC. XG performed the computational analysis. JC, RM, Zhifu Han and JN performed data analysis and contributed to manuscript preparation. JC wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
Takayama S, Isogai A . Self-incompatibility in plants. Annu Rev Plant Biol 2005; 56:467–489.
Kitashiba H, Nasrallah JB . Self-incompatibility in Brassicaceae crops: lessons for interspecific incompatibility. Breed Sci 2014; 64:23–37.
Stein JC, Howlett B, Boyes DC, Nasrallah ME, Nasrallah JB . Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc Natl Acad Sci USA 1991; 88:8816–8820.
Schopfer CR, Nasrallah ME, Nasrallah JB . The male determinant of self-incompatibility in Brassica. Science 1999; 286:1697–1700.
Shiba H, Takayama S, Iwano M, et al. A pollen coat protein, SP11/SCR, determines the pollen S-specificity in the self-incompatibility of Brassica species. Plant Physiol 2001; 125:2095–2103.
Suzuki G, Kai N, Hirose T, et al. Genomic organization of the S locus: identification and characterization of genes in SLG/SRK region of S(9) haplotype of Brassica campestris (syn. rapa). Genetics 1999; 153:391–400.
Nasrallah JB, Kao TH, Chen CH, Goldberg ML, Nasrallah ME . Amino-acid sequence of glycoproteins encoded by three alleles of the S locus of Brassica oleracea. Nature 1987; 326:617–619.
Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, Hinata K . The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 2000; 403:913–916.
Kachroo A, Schopfer CR, Nasrallah ME, Nasrallah JB . Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 2001; 293:1824–1826.
Takayama S, Shimosato H, Shiba H, et al. Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 2001; 413:534–538.
Takayama S, Shiba H, Iwano M, et al. The pollen determinant of self-incompatibility in Brassica campestris. Proc Natl Acad Sci USA 2000; 97:1920–1925.
Ivanov R, Fobis-Loisy I, Gaude T . When no means no: guide to Brassicaceae self-incompatibility. Trends Plant Sci 2010; 15:387–394.
Shimosato H, Yokota N, Shiba H, et al. Characterization of the SP11/SCR high-affinity binding site involved in self/nonself recognition in Brassica self-incompatibility. Plant Cell 2007; 19:107–117.
Iwano M, Takayama S . Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol 2012; 15:78–83.
Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH . Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004; 16:1220–1234.
Shiu SH, Bleecker AB . Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE 2001; 2001:re22.
Naithani S, Chookajorn T, Ripoll DR, Nasrallah JB . Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc Natl Acad Sci USA 2007; 104:12211–12216.
Kusaba M, Nishio T, Satta Y, Hinata K, Ockendon D . Striking sequence similarity in inter- and intra-specific comparisons of class I SLG alleles from Brassica oleracea and Brassica campestris: implications for the evolution and recognition mechanism. Proc Natl Acad Sci USA 1997; 94:7673–7678.
Sato K, Nishio T, Kimura R, et al. Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics 2002; 162:931–940.
Kemp BP, Doughty J . S cysteine-rich (SCR) binding domain analysis of the Brassica self-incompatibility S-locus receptor kinase. New Phytol 2007; 175:619–629.
Boggs NA, Dwyer KG, Nasrallah ME, Nasrallah JB . In vivo detection of residues required for ligand-selective activation of the S-locus receptor in Arabidopsis. Curr Biol 2009; 19:786–791.
Mishima M, Takayama S, Sasaki K, et al. Structure of the male determinant factor for Brassica self-incompatibility. J Biol Chem 2003; 278:36389–36395.
Chookajorn T, Kachroo A, Ripoll DR, Clark AG, Nasrallah JB . Specificity determinants and diversification of the Brassica self-incompatibility pollen ligand. Proc Natl Acad Sci USA 2004; 101:911–917.
Sauerborn MK, Wright LM, Reynolds CD, Grossmann JG, Rizkallah PJ . Insights into carbohydrate recognition by Narcissus pseudonarcissus lectin: the crystal structure at 2 A resolution in complex with alpha1-3 mannobiose. J Mol Biol 1999; 290:185–199.
Wouters MA, Rigoutsos I, Chu CK, Feng LL, Sparrow DB, Dunwoodie SL . Evolution of distinct EGF domains with specific functions. Protein Sci 2005; 14:1091–1103.
Cooke RM, Wilkinson AJ, Baron M, et al. The solution structure of human epidermal growth factor. Nature 1987; 327:339–341.
Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF . Met, metastasis, motility and more. Nat Rev Mol Cell Biol 2003; 4:915–925.
Almeida MS, Cabral KM, Kurtenbach E, Almeida FC, Valente AP . Solution structure of Pisum sativum defensin 1 by high resolution NMR: plant defensins, identical backbone with different mechanisms of action. J Mol Biol 2002; 315:749–757.
He XL, Li HM, Zeng ZH, Liu XQ, Wang M, Wang DC . Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel. J Mol Biol 1999; 292:125–135.
Giranton JL, Dumas C, Cock JM, Gaude T . The integral membrane S-locus receptor kinase of Brassica has serine/threonine kinase activity in a membranous environment and spontaneously forms oligomers in planta. Proc Natl Acad Sci USA 2000; 97:3759–3764.
Lemmon MA, Schlessinger J . Cell signaling by receptor tyrosine kinases. Cell 2010; 141:1117–1134.
Han Z, Sun Y, Chai J . Structural insight into the activation of plant receptor kinases. Curr Opin Plant Biol 2014; 20:55–63.
Liu T, Liu Z, Song C, et al. Chitin-induced dimerization activates a plant immune receptor. Science 2012; 336:1160–1164.
Sato Y, Okamoto S, Nishio T . Diversification and alteration of recognition specificity of the pollen ligand SP11/SCR in self-incompatibility of Brassica and Raphanus. Plant Cell 2004; 16:3230–3241.
Otwinowski Z, Minor W . Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 1997; 276:307–326.
Sheldrick GM . A short history of SHELX. Acta Crystallogr A 2008; 64:112–122.
Emsley P, Cowtan K . Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126–2132.
Adams PD, Grosse-Kunstleve RW, Hung LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 2002; 58:1948–1954.
Xiang Z, Soto C, Honig B . Evaluating conformational free energies: the colony energy and its application to the problem of loop prediction. Proc Natl Acad Sci USA 2002; 99:7432–7437.
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y . The I-TASSER Suite: protein structure and function prediction. Nat Methods 2015; 12:7–8.
Gong X, Wang P, Yang F, et al. Protein-protein docking with binding site patch prediction and network-based terms enhanced combinatorial scoring. Proteins 2010; 78:3150–3155.
Pronk S, Pail S, Schulz R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013; 29:845–854.
Acknowledgements
We thank J He at Shanghai Synchrotron Radiation Facility (SSRF) for assistance with x-ray data collection. This research was funded by grants from Projects of International Cooperation and Exchanges NSFC (31420103906), National Natural Science Foundation of China (31130063 and 31421001), and Chinese Ministry of Science and Technology (2015CB910200) to JC.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Table S1
Data collection and Refinement Statistics (PDF 259 kb)
Supplementary information, Figure S1
Structure of the two N-terminal lectin domains of SRK9. (PDF 109 kb)
Supplementary information, Figure S2
Sequence alignment of the N-terminal lectin domains of eSRK9 with S-domain proteins from Arabidopsis thaliana. (PDF 773 kb)
Supplementary information, Figure S3
Conserved structures of SCRs from Brassica rapa. (PDF 203 kb)
Supplementary information, Figure S4
Sequence alignment of eSRKs from Brassica rapa. (PDF 714 kb)
Supplementary information, Figure S5
Residues mediating eSRK9 homodimerization. (PDF 72 kb)
Supplementary information, Figure S6
Sequence alignment of eSRK9 with SLG9. (PDF 168 kb)
Supplementary information, Figure S7
Modeling studies of additional self and non-self eSRK-SCR pairs. (PDF 257 kb)
Rights and permissions
About this article
Cite this article
Ma, R., Han, Z., Hu, Z. et al. Structural basis for specific self-incompatibility response in Brassica. Cell Res 26, 1320–1329 (2016). https://doi.org/10.1038/cr.2016.129
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2016.129
Keywords
This article is cited by
-
Self-incompatibility: a targeted, unexplored pre-fertilization barrier in flower crops of Asteraceae
Journal of Plant Research (2023)
-
Ancestral self-compatibility facilitates the establishment of allopolyploids in Brassicaceae
Plant Reproduction (2023)
-
A pair of non-Mendelian genes at the Ga2 locus confer unilateral cross-incompatibility in maize
Nature Communications (2022)
-
Mechanism of self/nonself-discrimination in Brassica self-incompatibility
Nature Communications (2020)
-
Aminooxyacetic acid (АОА), inhibitor of 1-aminocyclopropane-1-carboxilic acid (AСС) synthesis, suppresses self-incompatibility-induced programmed cell death in self-incompatible Petunia hybrida L. pollen tubes
Protoplasma (2020)