Abstract
Neuroblastoma (NBL) is the most common solid tumor in infants and accounts for 15% of all pediatric cancer deaths. Several risk factors predict NBL outcome: age at the time of diagnosis, stage, chromosome alterations and MYCN (V-Myc Avian Myelocytomatosis Viral Oncogene Neuroblastoma-Derived Homolog) amplification, which characterizes the subset of the most aggressive NBLs with an overall survival below 30%. MYCN-amplified tumors develop exceptional chemoresistance and metastatic capacity. These properties have been linked to defects in the apoptotic machinery, either by silencing components of the extrinsic apoptotic pathway (e.g. caspase-8) or by overexpression of antiapoptotic regulators (e.g. Bcl-2, Mcl-1 or FLIP). Very little is known on the implication of death receptors and their antagonists in NBL. In this work, the expression levels of several death receptor antagonists were analyzed in multiple human NBL data sets. We report that Lifeguard (LFG/FAIM2 (Fas apoptosis inhibitory molecule 2)/NMP35) is downregulated in the most aggressive and undifferentiated tumors. Intringuingly, although LFG has been initially characterized as an antiapoptotic protein, we have found a new association with NBL differentiation. Moreover, LFG repression resulted in reduced cell adhesion, increased sphere growth and enhanced migration, thus conferring a higher metastatic capacity to NBL cells. Furthermore, LFG expression was found to be directly repressed by MYCN at the transcriptional level. Our data, which support a new functional role for a hitherto undiscovered MYCN target, provide a new link between MYCN overexpression and increased NBL metastatic properties.
Similar content being viewed by others
Main
Neuroblastoma (NBL) is the most common solid tumor of infancy, accounting for 15% of all pediatric cancer deaths. These tumors are very heterogeneous, with a clinical course ranging from spontaneous regression to aggressive behavior. Several risk factors predict NBL outcome: INSS (International Neuroblastoma Staging System) tumor stage, age at diagnosis, INPC (International Neuroblastoma Pathology Classification) classification, DNA ploidy and MYCN (V-Myc Avian Myelocytomatosis Viral Oncogene Neuroblastoma-Derived Homolog) oncogene amplification, which characterizes the subset of most aggressive NBLs with overall survival below 30%.1, 2, 3 MYCN-amplified tumors are characterized by exceptional chemoresistance and metastatic capacity. These properties have been linked to defects in the apoptotic arsenal, either by overexpression of the antiapoptotic regulators of the mitochondrial pathway (e.g. Bcl-2, Mcl-1)4, 5 or by alteration of components of the extrinsic apoptotic pathway (e.g. caspase-8).6, 7, 8 In fact, there is evidence that the extrinsic pathway may serve as a checkpoint to guard cells from MYCN-initiated tumorigenesis as MYCN-elevated levels sensitize NBL cells to death receptor (DR)-induced cell death, either by TRAIL, TNFα or FasL stimuli.9 To date, the main mechanism underlying the lack of DR-induced apoptosis in MYCN-amplified tumors has been methylation of the caspase-8 promoter, which blocks its expression and renders cells resistant to DR-induced cell death.7, 10 However, the correlation between MYCN amplification and caspase-8 silencing in tumor samples remains controversial; other authors showed the inactivation of caspase-8 to be independent of MYCN amplification and NBL prognosis.6 As MYCN amplification and caspase-8 silencing may not occur simultaneously, alternative resistance mechanisms must exist, which either block DR-induced cell death or switch DR signaling to alternative functions. DR activity can also be modulated by DR antagonists, which have been poorly characterized in the context of NBLs. The present work sought to analyze the role of DR antagonists in NBL and their contribution to the oncogenic properties of NBL cells. Several DR antagonists were found to be differentially expressed in the highest-risk NBL tumors, namely stage 4 MYCN-amplified NBLs. Among these, FAIM2 (Fas apoptosis inhibitory molecule 2), most commonly referred to as Lifeguard (LFG), correlates best with worse overall survival of NBL patients. Furthermore, we show that MYCN is able to repress directly LFG expression, which results in increased oncogenic properties such as augmented sphere formation, decreased adhesion and enhanced migration. Collectively, our results demonstrate a previously unappreciated role of LFG, and support a new target for therapeutic intervention against high-risk NBL.
Results
The DR antagonist LFG is downregulated in high-risk NBL
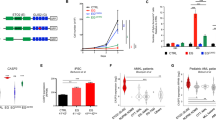
The mRNA expression levels of antiapoptotic genes known to modulate the extrinsic pathway in the neural lineage were analyzed in independent human expression and prognostic NBL data sets. Few DR modulators were consistently altered in different NBL data sets. Table 1 shows the fold change variation between stage 4 MYCN-amplified tumors versus the rest of stages present in the respective study (i.e. stages 1, 3 and 4 without MYCN amplification). PEA15, CFLAR/FLIP and LFG were downregulated in advanced stages of the disease. In particular, the highest differential expression was found in stage 4 MYCN-amplified patients, the group of NBLs with the worst prognosis (Figure 1a). Whether the differences in gene expression could be associated with patient outcome was also analyzed. Notably, only low expression levels of LFG were associated with worse overall survival (Figure 1b and Table 1). Moreover, the association between LFG levels and patient survival was maintained in MYCN non-amplified tumor subsets (Supplementary Figure 1). These differences in LFG expression levels were then validated in an independent cohort of 38 primary tumor samples obtained at the Vall d’Hebron Hospital (Supplementary Table 1). Quantitative real-time PCR (qPCR) expression analysis revealed a significant reduction in LFG levels in stage 4 and MYCN-amplified tumors (Figure 1c). Western blot analyses confirmed that differences in mRNA expression were also found at the protein level (Figure 1d). In summary, we conclude that LFG is the DR antagonist with the highest differential expression and association with patient outcome in high-risk NBL.
LFG is downregulated in high-risk NBL. (a) mRNA expression of the DR antagonists FLIP (1868_g_at), PEA15 (32260_at) and LFG (33293_at) in different NBL stages obtained from GSE3960 data set. Data were analyzed by one-way ANOVA; *P<0.05, **P<0.01 and ***P<0.001. MNA, MYCN amplified. (b) Kaplan–Meier overall survival curve in 251 tumors based on high or low LFG expression (probe A_23_P139891). (c) Relative LFG expression measured by qPCR in an independent cohort of NBL primary tumors collected at Vall d’Hebron Hospital. Data were analyzed by one-way ANOVA; *P<0.05. (d) Western blot showing LFG expression in MYCN non-amplified versus four MYCN-amplified primary NBL tumors. Naphtol blue (NB) staining was used as a loading control
LFG is associated with NBL differentiation
When LFG levels were correlated with a number of clinicopathologic variables (i.e. age at diagnosis, site of primary tumor, site of metastasis and histology), LFG expression was found to be closely associated with tumors presenting unfavorable histology (P=0.0041) and MYCN amplification (P=0.0121), which are often undifferentiated tumors11 (Figure 2a and Supplementary Table 2). With a view to ascertain the association between tumor differentiation and LFG expression, we sought to characterize LFG levels in the different status of NBL cell differentiation. On the one hand, all-trans retinoic acid (RA)-mediated differentiation of NBL cell lines increased LFG at mRNA and protein levels (Figures 2b and c). TRKB was used as a neuronal differentiation marker12 to confirm the differentiation response of the NBL cell lines to the RA treatment. On the other hand, LFG levels were analyzed in cells cultured in neurosphere formation medium (serum-free and supplemented with EGF/FGF2), which mimics undifferentiated status of NBL cells.13, 14 When NBL cell lines were seeded in neurosphere medium, cells formed non-adherent spheres. LFG was found to be downregulated in sphere medium-cultured cells compared with cells cultured in standard conditions (P=0.0286 for SK-N-BE(2) and IMR-5; P=0.0294 for IMR-32) (Figure 2d). We conclude that LFG expression associates with the differentiation status of NBL cells.
LFG levels are associated with NBL differentiation. (a) Relative LFG expression in histologically favorable (n=30) versus unfavorable (n=8) primary tumors. mRNA levels were measured by qPCR. **P<0.01, unpaired t-test. (b) All-trans RA (10 μM) induced differentiation in NBL cell lines SK-N-BE(2) and SH-SY5Y. LFG and TRKB expression were measured by qPCR at the indicated time points. Data are presented as mean±S.E.M. of three independent experiments. (c) Western blot of LFG expression. α-Tubulin was used as a loading control. Bands were quantified using Image J software (National Institutes of Health, Bethesda, MD, USA) and normalized expression values are indicated. (d) LFG expression detection by qPCR in the indicated NBL cell lines cultured with standard conditions (Adherent) or using neurosphere medium (Sphere). *P<0.05, unpaired t-test; data are represented as mean±S.E.M., n=4. Images (right) show IMR-5 adherent and sphere-forming cultures. Scale bar, 100 μm
Reduction of LFG levels modulates proliferation, adhesion and migration of NBL cells
To assess the role of LFG in NBL oncogenic properties, loss-of-function experiments were performed using two different short hairpin (sh)RNA targeting LFG (Figure 3a). Both shLFG constructs notably knocked down LFG at the protein level and reduced proliferation of SH-SY5Y (P=0.022 for shLFG and P=0.0238 for shLFG2) and CHLA-90 cells (P=0.0043 for shLFG and P=0.022 for shLFG2) (Figure 3b). The most efficient downregulation was achieved by shLFG, which was used for the following experiments. Cell adhesion assays showed that shLFG-transduced cells exhibit around 50% reduction in adhesion to adherent plates compared with scrambled (Scr)-infected cells (P=0.0286 for SH-SY5Y and P=0.0043 for CHLA-90 cells) (Figure 3c). Moreover, when NBL cells were seeded at low density, loss of LFG increased non-adherent growth compared with the Scr shRNA (Figure 3d). More than 10% of shLFG cells were able to form viable spheres when they were seeded at low density in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% fetal bovine serum (FBS), whereas <4% of control cells did so (P=0.0066). Furthermore, shLFG-infected cells showed a fourfold increased migration through the 8 μm-transwell polycarbonate inserts (P=0.0191) (Figure 3e). The impact of these properties was further evaluated in vivo. SH-SY5Y cells (shLFG or shScr) were injected into the flank of female NMRI-Foxn1nu/Foxn1nu mice. Tumors appeared earlier in the LFG knockdown group (Figure 4a). No significant differences in tumor growth or tumor weight were found (Figures 4b and c). The presence of spontaneous metastasis was evaluated in the lungs by ex vivo bioluminiscence (BLI) detection. Mice inoculated with LFG knockdown cells showed increased metastatic burden in the lungs and higher number of metastatic lesions (Figures 4d and e). Taken together, these results indicate that the reduction on LFG levels may contribute to altered adhesion properties and increased metastatic potential of NBL cells.
LFG knockdown favors the metastatic potential of NBL cell lines. (a) Western blot showing LFG downregulation after infection with shScr or shLFG lentiviruses using anti-FAIM2 (H-21) antibody (Santa Cruz Biotechnology Inc). Bands were quantified and normalized versus α-tubulin. (b) Cell proliferation assay in SH-SY5Y and CHLA-90 cells after LFG downregulation with two shRNA sets (shScr/shLFG and shScr2/shLFG2). Cells were fixed with 4% paraformaldehyde at the indicated time points. Data are presented as mean±S.E.M. of three independent experiments. *P<0.05 and **P<0.01, unpaired t-test; * shScr versus shLFG; #shScr2 versus shLFG2. (c) Cell adhesion assay of SH-SY5Y and CHLA-90 transduced with either Scr or LFG shRNAs, measured at the indicated time points. *P<0.05 and **P<0.01, unpaired t-test; data are represented as mean±S.E.M., n=3. (d) Sphere formation assay of shScr and shLFG SH-SY5Y cells seeded at low density in DMEM+15% FBS and kept in standard culture conditions for 4 weeks. Graph represents the percentage of sphere formation. **P<0.01, unpaired t-test; data are presented as mean±S.E.M., n=9. Infection efficiency was monitored by visualization of green fluorescent protein (GFP). Scale bar, 100 μm. (e) Transwell migration assay of shScr and shLFG-transduced SH-SY5Y cells. Cells that migrated were quantified by WST-1 assay. *P<0.05, unpaired t-test; graphs show mean±S.E.M., n=3
LFG knockdown increases the metastatic potential of SH-SY5Y cells. (a) Xenograft formation assay with shScr and shLFG SH-SY5Y cells injected into the flank of NMRI nude mice. (b) Tumor volume measured at different days postinjection using a digital caliper. Data are presented as mean±S.E.M. (c) Tumor weight measured after surgery. Data are presented as mean±S.E.M. (d) Total BLI flux (photons/s) of lung metastases from shScr and shLFG SH-SY5Y-injected mice. Images are representative of ex vivo BLI lung images. Pictures were set at the same scale to compare BLI between shScr and shLFG conditions. (e) Scoring of lung metastases. Data are shown as mean±S.E.M. Images are representative of ex vivo BLI showing distant metastases in lungs disected from shScr and shLFG-injected mice
LFG knockdown alters cell adhesion programs
To enlighten the mechanism by which LFG knockdown reduces cell adhesion and enhances migration properties of NBL cells, we proceeded to LFG knockdown and performed a whole transcriptome analysis by Affymetrix expression arrays. Principal component analysis segregated the infected cells (Scr versus shLFG), indicating a consistent transcriptional impact of LFG knockdown (Supplementary Figure 2). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis15, 16 of genes differentially expressed in response to LFG knockdown revealed a reproducible modulation of genes mainly involved in cancer pathways and adhesion processes (Figure 5a). Functional annotation analysis showed a remarkable modulation of genes implicated in focal adhesion, cell adhesion and adherent junctions (Figure 5b), which supported an active role of LFG in the tumorigenic properties of NBL cells. Array data were validated by qPCR for FGF14, STK36, BCL2, RAP1A, TJP1, ITGA9 and LFG (Figure 5c).
LFG knockdown alters the expression of cell adhesion molecules. (a) KEGG pathway enrichment analysis of differentially expressed genes between shScr and shLFG-transduced SH-SY5Y cells. (b) Gene set enrichment analysis performed on genes differentially expressed between shScr and shLFG SH-SY5Y cells. Heatmap shows selected differentially expressed genes grouped in categories of functionally relevant pathways. (c) Array validation. qPCR using primers for three upregulated genes (FGF14, STK36 and BCL2) and three downregulated genes (RAP1A, TJP1 and ITGA9). LFG levels have been included as a control of the experiment. Values are represented as fold change versus shScr and are the mean±S.E.M. of three independent experimets
LFG expression is directly modulated by MYCN
As LFG levels were significantly lower in MYCN-amplified tumors, we sought to determine whether MYCN participates in LFG repression. Our first approach consisted in using the SHEP MYCN-inducible Tet21N cell line, which constitutively overexpresses MYCN under the control of a tetracycline-responsive repressor element.17 Doxycycline treatment of Tet21N resulted in a differentiated phenotype (Figure 6a) and reduced MYCN expression with a concomitant time-dependent upregulation of LFG, both at the mRNA and protein level (Figure 6b). Furthermore, the same result was obtained when MYCN was silenced using an inducible shRNA in an independent MYCN-amplified cell line such as IMR-32 (Supplementary Figure 4).
MYCN represses directly LFG transcription. (a) Representative images of MYCN-Tet-off-inducible Tet21N cells treated with or without doxycycline (100 ng/ml). Scale bar, 100 μm. (b) Tet21N cell line treated with doxycycline (100 ng/ml) for the indicated times. LFG and MYCN mRNA levels were detected by qPCR and normalized versus L27 (upper panel) and also detected by western blot (lower panel). Data on the graph are presented as mean±S.E.M., n=3. α-Tubulin was used as a loading control. Bands were quantified and normalized versus α-tubulin. (c) Identification of MYCN binding site at the LFG promoter (gene ID: FAIM2) by ChIP in Tet21N cells. The scale on the top of the panel indicates position on chromosome 12. Green bars represent fluorescent intensity of probes around the FAIM2 promoter, expressed as log 2 ratios. The red bar represents a high confidence MYCN peak, identified by the NimbleScan peak finding algorithm. (d) LFG levels measured by qPCR in the indicated cell lines treated with 1 μM of the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine or 1 mM of the HDAC inhibitor sodium butyrate or for the indicated times. Mean±S.E.M., n=3
To address whether MYCN represses LFG directly, a chromatin immunoprecipitation (ChIP) data set was examined for MYCN binding in Tet21N cells with and without doxycycline treatment.18 A high confidence MYCN-binding peak was identified in the LFG proximal promoter that disappeared when MYCN expression was repressed by doxycycline treatment (Figure 6c).
MYCN-mediated transcription repression is now considered to be at least as important as transcription activation.19 However, the repressing mechanisms remain unclear. It has been proposed that MYCN may interact with transcription factors or transcriptional complexes, inducing DNA and histone modifications. To further characterize how MYCN represses LFG transcription, MYCN-amplified NBL cell lines were treated with the the DNA methyltransferase inhibitor 5-aza-2'-deoxycytidine and the histone deacetylase (HDAC) inhibitor sodium butyrate (Figure 6d). Both treatments induced an increase in LFG mRNA levels in all cell lines tested, thereby suggesting that MYCN represses LFG expression by favoring histone deacetylation and LFG DNA promoter methylation.
Discussion
Alterations of key elements of the apoptotic pathway have been shown to be determinant in the outcome of NBLs. While characterization of the mitochondrial or intrinsic pathway elements has been extensive, limited attention to the components of the extrinsic or DR pathway has been paid. The most characterized element of the extrinsic pathway is caspase-8, which is lost in ∼70% of NBLs.6, 20 Loss of caspase-8 has been shown to prevent DR-induced apoptosis21, 22, 23 and potentiate NBL metastasis.24, 25 However, caspase-8 can also be prometastatic if other components of the apoptotic machinery are compromised.26 Whether other modulators of the intrinsic pathway can impact similarly to caspase-8 in NBL behavior remain understudied.
Here, we have analyzed the expression of several DR antagonists in multiple NBL data sets and found that LFG was the most differentially expressed DR antagonist in MYCN-amplified tumors. LFG low levels were also found to correlate with worse overall survival. LFG was originally reported to be expressed exclusively in the nervous system and upregulated during the development of rat sciatic nerve.27 Primary functional analyses revealed that LFG is able to antagonize FasL but not TNFα-induced cell death in T cells.28 We and others confirmed that LFG is essential to prevent Fas-induced apoptosis in neuronal cells29 by clustering with Fas receptor and blocking caspase-8 activation.30 Recently, an alternative LFG function has been proposed. Merianda and co-workers31 have demonstrated that LFG localization in axons is sufficient to support axonal growth in dorsal root ganglionar cells, thereby indicating that LFG may have DR antagonist-independent functions. Alternative functions for other DR antagonists are rare but not unique. In fact, the antiapoptotic protein PEA15 has also been linked to NBL prognosis. High levels of this protein correlate with favorable clinical features. PEA15 was shown to block NBL migration through the inhibition of ERK/RSK2 signaling, suggesting a metastatic limiting role in the progression of NBL.32 Similar to PEA15, loss of LFG potentiates cell detachment, adherence-independent growth, migration and metastasis. However, the mechanism seems to be quite different. LFG knockdown impacted indirectly the modulation of cell adhesion and some epithelial-to-mesenchymal (EMT) genes (Supplementary Figure 3). One example is ITGA9, which is found to be downregulated in shLFG-infected cells. ITGA9 has been shown to be essential for the adhesion of rhabdomyosarcoma cells.33 Other prometastatic integrins were found to be upregulated (e.g. ITGAV, ITGB1). For example, overexpression of ITGAV has been found to correlate with metastasis of laryngeal,34 hypopharyngeal squamous cell carcinomas35 and colorectal cancer.36 Moreover, ITGAV knockdown decreased proliferation and increased apoptosis in Hep-2 carcinoma cells.35 Several tight-junction (TJ) genes were also downregulated (e.g. TJP1). Loss of cohesion of the TJ structure can lead to invasion and ultimately to the metastasis of cancer cells.37 Regarding the EMT-related genes, we have found upregulation of TWIST1 upon LFG knockdown and also an inverse correlation in human samples (Supplementary Figures 3a and b). TWIST1 is an important EMT and metastasis promoter gene,38 which has been recently found to to be overexpressed in MYCN-amplified NBL.39 By contrast, ERBB3 and KRT19 genes were found to be reduced in NBL cells infected with shLFG and downregulated in NBL human samples with low levels of LFG. ERBB3 belongs to the epidermal growth factor receptor family and participates in the activation of proliferation40 and differentiation pathways,41 whereas KRT19 is a member of the keratin family and participates in the organization of the cytoskeleton.42 The expression of KRT19 or ERBB3 was found to be low in patients with poor prognosis.39
Other genes such as FGF14, STK36 or BCL2 may contribute in a different manner to the metastatic process. STK36 is a serine/threonine kinase that has an important role in the activation of the sonic hedgehog pathway, reported to be essential for NBL proliferation and tumor growth.43 The upregulation of BCL2 may also contribute to inhibit apoptosis when cells grow in anchorage-independent conditions.
Furthermore, we provide evidence that low expression of LFG is not a mere association with the differentiation status of the cell, but it is actively repressed by the MYCN oncogene. MYCN encodes a protein with a basic helix–loop–helix domain known to be a transcriptional regulator of genes controlling cell cycle and proliferation, cell invasion, angiogenesis and cell survival and death pathways (reviewed in Westermark et al.11). MYCN has been shown to participate in NBL metastasis by direct or indirect repression of integrin subunits altering cell–matrix or cell–cell interactions44 or repressing Caveolin-1, whose downregulation elicits anchorage-independent growth and tumor formation.45 We show that repression of LFG impacts on the expression of several molecules that participate in cell adhesion processes, recapitulating some of the MYCN reported effects.
In summary, our results provide a new link between MYCN amplification and early steps of NBL metastasis, and the participation of LFG in unprecedented functions such as cell adhesion, anchorage-independent growth and migration.
Materials and Methods
Reagents
Unless otherwise indicated, all reagents were purchased from Sigma-Aldrich (Barcelona, Spain). 5-Aza-2'-deoxycytidine was purchased from Calbiochem (San Diego, CA, USA).
Analysis of mRNA NBL data sets
Gene expression data of 101 and 51 primary NBL tumors (GSE3960;46 GSE1623747) were used to analyze the expression of DR antagonists. The expression of each individual DR antagonist was analyzed comparing the stage 4 MYCN-amplified tumors versus the other stages present in the study (i.e. stages 1, 3 and 4 without MYCN amplification). Gene expression data of human samples (GSE1647648 and Oberthuer lab data set from Oncogenomics database) were used to generate Kaplan–Meier survival analysis.
Human samples
Primary tumor tissue samples from 38 NBL patients enrolled at the Vall d’Hebron Hospital (Barcelona, Spain) were obtained immediately after surgery and snap frozen in liquid nitrogen and stored at −80 °C until processing. Tumors were examined by the pathologist to confirm NBL diagnosis and the presence of at least 80% of tumor tissue sample and histopathologic classification. All patients gave written informed consent.
Gene expression analysis by qPCR
Total RNA was extracted from NBL cell lines and tumor samples using the RNeasy Mini Kit (Qiagen, Las Matas, Spain). Between 0.5 and 1 μg of total RNA was retrotranscribed using High Capacity RNA-to-cDNA Kit (Applied Biosystems, Alcobendas, Spain). Gene expression analyses were performed using the LFG, TRKB, FGF14, STK36, BCL2, RAP1A, TJP1 and ITGA9 Taqman probes. Unless otherwise indicated, all expression analysis were normalized versus 18S housekeeping gene. MYCN expression was performed with SYBR green (Applied Biosystems) and normalized against L27 using the following primers: MYCN – forward, 5′-CCTGAGCGATTCAGATGATGA-3′ and reverse, 5′-GGTGAATGTGGTGACAGCCT-3′; L27 – forward, 5′-AGCTGTCATCGTGAAGAA-3′ and reverse, 5′-CTTGGCGATCTTCTTCTTGCC-3′. Analysis was performed using the 7900HT Sequence Detection Systems 2.3 Software (Applied Biosystems). The relative fold change in expression was determined by the comparative 2(−ΔΔCT) method.49
Western blot
Cell homogenates were obtained in lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton) and tissue homogenates were obtained in RIPA buffer (Pierce/ThermoFisher Scientific, Lafayette, CO, USA), both supplemented with 1 × EDTA-free complete protease inhibitor cocktail (Roche, Sant Cugat del Vallès, Spain). Protein concentration was quantified by a modified Lowry assay (DC protein assay; Bio-Rad, Hercules, CA, USA). Proteins (20–30 μg) were resolved in Tris/glycine SDS/PAGE gels and transferred to PVDF membranes (Millipore, Billerica, MA, USA). After blocking with Tris-buffered saline with Tween-20 containing 5% nonfat dry milk for 1 h at room temperature, membranes were probed with anti-LFG (AnaSpec, Seraing, Belgium) or anti-FAIM2 (H-21) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) when indicated, and anti-α-tubulin or anti-MYCN NCM-II-100 (Abcam, Cambridge, UK) primary antibodies overnight at 4 °C. Membranes were developed with SuperSignal Dura Detection Kit (Pierce/ThermoFisher Scientific) or EZ-ECL Chemiluminescence Detection Kit (Biological Industries, Kibbutz Beit-Haemek, Israel).
Cell lines
SH-SY5Y, IMR-32 and IMR-5 were acquired from the American Type Tissue Collection (ATCC, Barcelona, Spain). CHLA-90 cells were obtained from Children’s Oncology Group Cell Line and Xenograft Repository (Lubbock, TX, USA). SK-N-BE(2) and LAI-5S cells were purchased from Public Health England Culture Collections (Salisbury, UK). Tet21N cells were a generous gift from Manfred Schwab (DKFZ, Heidelberg, Germany). IMR-32 cells transduced with the conditional MYCN shRNA were kindkly provided by Dr Frank Westermann (DKFZ, Heidelberg, Germany). All cell lines obtained directly from the tissue banks were amplified and stored in liquid nitrogen. Upon resuscitation, cells were maintained in culture for no more than 2 months. SH-SY5Y, IMR-5, IMR-32 and LAI-5S cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 15% heat-inactivated FBS, 20 U/ml penicillin and 20 μg/ml streptomycin. SK-N-BE(2) and CHLA-90 were maintained in Iscove’s modified Dulbecco’s medium, supplemented with 20% heat-inactivated FBS, 1% of insulin-transferrin-selenium, 100 U/ml penicillin and 100 μg/ml streptomycin. Tet21N cells were grown in RPMI (Roswell Park Memorial Institute) 1640 medium (Invitrogen, Barcelona, Spain) supplemented with 10% heat-inactivated FBS, 25 mM HEPES (Invitrogen), 200 μg/ml geneticin (G418), 0.5 μg/ml amphotericin B, 10 μg/ml hygromycin B, 100 U/ml penicillin and 100 μg/ml streptomycin. All cultures were maintained at 37 °C in a saturated atmosphere of 95% air and 5% CO2.
RA differentiation
Induction of differentiation of NBL cell lines was performed as described previously by Encinas et al.12 Briefly, 6 × 105 cells were seeded in 100 mm culture dishes previously coated with 65 μg/ml type I collagen (BD Biosciences, San Agustin de Guadalix, Spain). All-trans RA was added the next day at a final concentration of 10 μM and maintained during 1, 3 and 5 days.
Sphere formation
SK-N-BE(2), IMR-5 and IMR-32 cells were grown for sphere formation in serum-free neurobasal medium (Invitrogen), supplemented with B27 (Invitrogen), 2 mM L-glutamine (Invitrogen), 20 ng/ml EGF (ProSpec-Tany Technogene Ltd, Ness-Ziona, Israel), 20 ng/ml FGF2 (ProSpec-Tany Technogene Ltd), 20 U/ml penicillin and 20 μg/ml streptomycin.
LFG knockdown
For loss-of-function experiments, two sets of shRNA lentiviral constructs were used: shScr and shLFG were previously described in Fernandez et al.30 and shScr2 and shLFG2 were purchased from ThermoFisher Scientific (RHS4531-EG23017). Lentiviruses were propagated using standard procedures.50, 51 NBL cells were seeded at 2 × 106 cells per 60 mm dish and infected with viral supernatant for 12 h. The transduction efficiency was evaluated 3 days later by scoring green fluorescent protein (GFP)-positive cells. LFG knockdown was assessed by qPCR and western blot.
Cell proliferation and adhesion assays
Infected NBL cells were seeded in 96-well plates (2 × 104 cells/well for proliferation assays and 2 × 105 cells/well for adhesion assays) and incubated at 37 °C for the indicated times. Then, cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Crystals were dissolved in 10% acetic acid and the absorbance was measured at 590 nm.
Migration assays
A suspension of 2.5 × 105 SH-SY5Y NBL cells was added in serum-free media to 8-μm-pore cell culture inserts (Falcon Discovery Labware; BD Biosciences). Cells were incubated for 8 h under standard culture conditions and complete media in the lower part of the chamber. Viable cells that had migrated to the lower transwell chamber were quantified by WST-1 assay (Roche).
Xenografts
A total of 5 × 106 SH-SY5Y-pFLuc cells infected with shScr (n=10) or shLFG (n=10) were injected in the flank of female NMRI nude mice in 300 μl of phosphate-buffered saline (PBS) and Matrigel (1 : 1). Tumor growth was measured every 2–3 days using an electronic caliper. At the respective scheduled surgery, 150 mg/kg D-Luciferin substrate (Promega Biotech Iberica, Madrid, Spain) was injected intraperitoneally 5 min before animals were killed. The primary tumor mass was excised and weighted. Lungs were removed to detect distant metastases. Each organ was immersed in a 300 μg/ml D-Luciferin solution in PBS and maintained in cold conditions until ex vivo BLI was performed using the IVIS Spectrum from the CIBER-BBN In Vivo Experimental Platform located at Vall d’Hebron Research Institute (VHIR).
mRNA microarray analysis
Expression profiling of triplicate experimental sample groups (shScr versus shLFG) was performed using Affymetrix microarray platform and the GeneChip Human Gene 1.0 ST Array (Affymetrix, Santa Clara, CA, USA). Two hundred nanograms of total RNA was hybridized to the arrays with the GeneChip WT Terminal Labeling and Hybridization Kit (Affymetrix). Chips were processed on an Affymetrix GeneChip Fluidics Station 450 and Scanner 3000 and normalization of the raw data (CEL files) was carried out with Robust Multichip Average algorithm.52 To filter and perform differential expression analyses, a moderated t-test (P<0.05 α-level) and fold-change thresholding (>33% reproducible change) were considered. The functional annotations of resulting gene lists were performed using the Gene Ontology53 and the KEGG15, 16 databases. To build the heatmaps, array expression values were normalized to the median and log 2 transformed. Values were converted to color scale using the MultipleExperimentViewer software (TM4 Software Suite, Boston, MA, USA54).
ChIP assays
Details of the MYCN chip experiments, along with validation studies, have been previously published by Murphy et al.18 Sites of enrichment were identified using the normalized log 2 ratios and the NimbleScan peak finding function.
Statistical analysis
Statistical significance was determined by unpaired t-test or one-way ANOVA when indicated using GraphPad (GraphPad Prism Software, La Jolla, CA, USA). *P<0.05, **P<0.01 and ***P<0.001.
Abbreviations
- BLI:
-
bioluminiscence
- ChIP:
-
chromatin immunoprecipitation
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- DR:
-
death receptor
- EMT:
-
epithelial-to-mesenchymal transition
- FAIM2:
-
Fas apoptosis inhibitory molecule 2
- FBS:
-
fetal bovine serum
- GFP:
-
green fluorescent protein
- HDAC:
-
histone deacetylase
- INPC:
-
International Neuroblastoma Pathology Classification
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LFG:
-
Lifeguard
- MYCN:
-
V-Myc Avian Myelocytomatosis Viral Oncogene Neuroblastoma-Derived Homolog
- NaB:
-
sodium butyrate
- NBL:
-
neuroblastoma
- PBS:
-
phosphate-buffered saline
- qPCR:
-
quantitative real-time polymerase chain reaction
- RA:
-
retinoic acid
- RPMI:
-
Roswell Park Memorial Institute
- Scr:
-
scrambled
- S.E.M.:
-
standard error of the mean
- Sh:
-
short hairpin
- TJ:
-
tight junction
References
Wagner LM, Danks MK . New therapeutic targets for the treatment of high-risk neuroblastoma. J Cell Biochem 2009; 107: 46–57.
Maris JM, Hogarty MD, Bagatell R, Cohn SL . Neuroblastoma. Lancet 2007; 369: 2106–2120.
Tonini GP, Nakagawara A, Berthold F . Towards a turning point of neuroblastoma therapy. Cancer Lett 2012; 326: 128–134.
Castle VP, Heidelberger KP, Bromberg J, Ou XG, Dole M, Nunez G . Expression of the apoptosis-suppressing protein Bcl-2, in neuroblastoma is associated with unfavorable histology and N-Myc amplification. Am J Pathol 1993; 143: 1543–1550.
Michels J, Johnson PWM, Packham G . Mcl-1. Int J Biochem Cell Biol 2005; 37: 267–271.
Fulda S, Poremba C, Berwanger B, Hacker S, Eilers M, Christiansen H et al. Loss of caspase-8 expression does not correlate with MYCN amplification, aggressive disease, or prognosis in neuroblastoma. Cancer Res 2006; 66: 10016–10023.
Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA et al. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med 2000; 6: 529–535.
Yang QW, Kiernan CM, Tian YF, Salwen HR, Chlenski A, Brumback BA et al. Methylation of CASP8, DCR2, and HIN-1 in neuroblastoma is associated with poor outcome. Clin Cancer Res 2007; 13: 3191–3197.
Cui HJ, Li T, Ding HF . Linking of N-Myc to death receptor machinery in neuroblastoma cells. J Biol Chem 2005; 280: 9474–9481.
van Noesel MM, Pieters R, Voute PA, Versteeg R . The N-myc paradox: N-myc overexpression in neuroblastomas is associated with sensitivity as well as resistance to apoptosis. Cancer Lett 2003; 197: 165–172.
Westermark UK, Wilhelm M, Frenzel A, Henriksson MA . The MYCN oncogene and differentiation in neuroblastoma. Semin Cancer Biol 2011; 21: 256–266.
Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, Cena V et al. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem 2000; 75: 991–1003.
Hansford LM, McKee AE, Zhang L, George RE, Gerstle JT, Thorner PS et al. Neuroblastoma cells isolated from bone marrow metastases contain a naturally enriched tumor-initiating cell. Cancer Res 2007; 67: 11234–11243.
Mahller YY, Williams JP, Baird WH, Mitton B, Grossheim J, Saeki Y et al. Neuroblastoma cell lines contain pluripotent tumor initiating cells that are susceptible to a targeted oncolytic virus. PLoS One 2009; 4: e4235.
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M . KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 2012; 40: D109–D114.
Kanehisa M, Goto S . KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000; 28: 27–30.
Lutz W, Stohr M, Schurmann J, Wenzel A, Lohr A, Schwab M . Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene 1996; 13: 803–812.
Murphy DM, Buckley PG, Bryan K, Das S, Alcock L, Foley NH et al. Global MYCN transcription factor binding analysis in neuroblastoma reveals association with distinct E-box motifs and regions of DNA hypermethylation. PLoS One 2009; 4: e8154.
Gherardi S, Valli E, Erriquez D, Perini G . MYCN-mediated transcriptional repression in neuroblastoma: the other side of the coin. Front Oncol 2013; 3: 42.
Grau E, Martinez F, Orellana C, Canete A, Yanez Y, Oltra S et al. Hypermethylation of apoptotic genes as independent prognostic factor in neuroblastoma disease. Mol Carcinogen 2011; 50: 153–162.
Hopkins-Donaldson S, Bodmer JL, Bourloud KB, Brognara CB, Tschopp J, Gross N . Loss of caspase-8 expression in highly malignant human neuroblastoma cells correlates with resistance to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res 2000; 60: 4315–4319.
Eggert A, Grotzer MA, Zuzak TJ, Wiewrodt BR, Ikegaki N, Brodeur GM . Resistance to TRAIL-induced apoptosis in neuroblastoma cells correlates with a loss of caspase-8 expression. Med Pediatr Oncol 2000; 35: 603–607.
Fulda S, Kufer MU, Meyer E, van Valen F, Dockhorn-Dworniczak B, Debatin KM . Sensitization for death receptor- or drug-induced apoptosis by re-expression of caspase-8 through demethylation or gene transfer. Oncogene 2001; 20: 5865–5877.
Stupack DG, Teitz T, Potter MD, Mikolon D, Houghton PJ, Kidd VJ et al. Potentiation of neuroblastoma metastasis by loss of caspase-8. Nature 2006; 439: 95–99.
Teitz T, Inoue M, Valentine MB, Zhu K, Rehg JE, Zhao W et al. Th-MYCN mice with caspase-8 deficiency develop advanced neuroblastoma with bone marrow metastasis. Cancer Res 2013; 73: 4086–4097.
Barbero S, Mielgo A, Torres V, Teitz T, Shields DJ, Mikolon D et al. Caspase-8 association with the focal adhesion complex promotes tumor cell migration and metastasis. Cancer Res 2009; 69: 3755–3763.
Schweitzer B, Taylor V, Welcher AA, McClelland M, Suter U . Neural membrane protein 35 (NMP35): a novel member of a gene family which is highly expressed in the adult nervous system. Mol Cell Neurosci 1998; 11: 260–273.
Somia NV, Schmitt MJ, Vetter DE, Van Antwerp D, Heinemann SF, Verma IM . LFG: an anti-apoptotic gene that provides protection from Fas-mediated cell death. Proc Natl Acad Sci USA 1999; 96: 12667–12672.
Beier CP, Wischhusen J, Gleichmann M, Gerhardt E, Pekanovic A, Krueger A et al. FasL (CD95L/APO-1L) resistance of neurons mediated by phosphatidylinositol 3-kinase-Akt/protein kinase B-dependent expression of lifeguard/neuronal membrane protein 35. J Neurosci 2005; 25: 6765–6774.
Fernandez M, Segura MF, Sole C, Colino A, Comella JX, Cena V . Lifeguard/neuronal membrane protein 35 regulates Fas ligand-mediated apoptosis in neurons via microdomain recruitment. J Neurochem 2007; 103: 190–203.
Merianda TT, Vuppalanchi D, Yoo S, Blesch A, Twiss JL . Axonal transport of neural membrane protein 35 mRNA increases axon growth. J Cell Sci 2012; 126 (Part 1): 90–102.
Gawecka JE, Geerts D, Koster J, Caliva MJ, Sulzmaier FJ, Opoku-Ansah J et al. PEA15 impairs cell migration and correlates with clinical features predicting good prognosis in neuroblastoma. Int J Cancer 2012; 131: 1556–1568.
Masia A, Almazan-Moga A, Velasco P, Reventos J, Toran N, Sanchez de Toledo J et al. Notch-mediated induction of N-cadherin and alpha9-integrin confers higher invasive phenotype on rhabdomyosarcoma cells. Br J Cancer 2012; 107: 1374–1383.
Lu JG, Li Y, Li L, Kan X . Overexpression of osteopontin and integrin alphav in laryngeal and hypopharyngeal carcinomas associated with differentiation and metastasis. J Cancer Res Clin Oncol 2011; 137: 1613–1618.
Lu JG, Sun YN, Wang C, Jin de J, Liu M . Role of the alpha v-integrin subunit in cell proliferation, apoptosis and tumor metastasis of laryngeal and hypopharyngeal squamous cell carcinomas: a clinical and in vitro investigation. Eur Arch Otorhinolaryngol 2009; 266: 89–96.
Viana Lde S, Affonso RJ Jr, Silva SR, Denadai MV, Matos D, Salinas de Souza C et al. Relationship between the expression of the extracellular matrix genes SPARC, SPP1, FN1, ITGA5 and ITGAV and clinicopathological parameters of tumor progression and colorectal cancer dissemination. Oncology 2013; 84: 81–91.
Martin TA, Mason MD, Jiang WG . Tight junctions in cancer metastasis. Front Biosci 2011; 16: 898–936.
Karreth F, Tuveson DA . Twist induces an epithelial–mesenchymal transition to facilitate tumor metastasis. Cancer Biol Ther 2004; 3: 1058–1059.
Nozato M, Kaneko S, Nakagawara A, Komuro H . Epithelial–mesenchymal transition-related gene expression as a new prognostic marker for neuroblastoma. Int J Oncol 2013; 42: 134–140.
Richards KN, Zweidler-McKay PA, Van Roy N, Speleman F, Trevino J, Zage PE et al. Signaling of ERBB receptor tyrosine kinases promotes neuroblastoma growth in vitro and in vivo. Cancer 2010; 116: 3233–3243.
Izycka-Swieszewska E, Wozniak A, Drozynska E, Kot J, Grajkowska W, Klepacka T et al. Expression and significance of HER family receptors in neuroblastic tumors. Clin Exp Metast 2011; 28: 271–282.
Zhou X, Liao J, Hu L, Feng L, Omary MB . Characterization of the major physiologic phosphorylation site of human keratin 19 and its role in filament organization. J Biol Chem 1999; 274: 12861–12866.
Xu L, Wang X, Wan J, Li T, Gong X, Zhang K et al. Sonic Hedgehog pathway is essential for neuroblastoma cell proliferation and tumor growth. Mol Cell Biochem 2012; 364: 235–241.
Judware R, Culp LA . Concomitant down-regulation of expression of integrin subunits by N-myc in human neuroblastoma cells: differential regulation of alpha2, alpha3 and beta1. Oncogene 1997; 14: 1341–1350.
Galbiati F, Volonte D, Engelman JA, Watanabe G, Burk R, Pestell RG et al. Targeted downregulation of caveolin-1 is sufficient to drive cell transformation and hyperactivate the p42/44 MAP kinase cascade. EMBO J 1998; 17: 6633–6648.
Wang Q, Diskin S, Rappaport E, Attiyeh E, Mosse Y, Shue D et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res 2006; 66: 6050–6062.
Ohtaki M, Otani K, Hiyama K, Kamei N, Satoh K, Hiyama E . A robust method for estimating gene expression states using Affymetrix microarray probe level data. BMC Bioinform 2010; 11: 183.
Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012; 483: 589–593.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001; 25: 402–408.
Naldini L, Blomer U, Gage FH, Trono D, Verma IM . Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA 1996; 93: 11382–11388.
Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol 1998; 72: 9873–9880.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249–264.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25: 25–29.
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003; 34: 374–378.
Acknowledgements
We thank members of the Vall d’Hebron Genomics Facility for array profiling, as well as the services of the VHIR Pathology, Animal Core Facilities and staff from the Molecular Imaging Platform. We are grateful to Dr. Frank Westermann (DKFZ, Heidelberg, Germany) for kindly providing us with the inducible shMYCN IMR-32 cell line. This work is supported by the Spanish Ministry of Science (MICINN) SAF2010-19953, CIBERNED CB06/05/1104, Generalitat de Catalunya SGR2009-346 and Instituto de Salud Carlos III (CP11/00052, RD12/0036/0016) cofinanced by the European Regional Development Fund (ERDF). LPF is supported by a FPU fellowship from the Spanish Ministry of Science.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Raschellà
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Planells-Ferrer, L., Urresti, J., Soriano, A. et al. MYCN repression of Lifeguard/FAIM2 enhances neuroblastoma aggressiveness. Cell Death Dis 5, e1401 (2014). https://doi.org/10.1038/cddis.2014.356
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.356