Abstract
Accurate methods to measure autophagic activity in vivo in neurons are not available, and most of the studies are based on correlative and static measurements of autophagy markers, leading to conflicting interpretations. Autophagy is an essential homeostatic process involved in the degradation of diverse cellular components including organelles and protein aggregates. Autophagy impairment is emerging as a relevant factor driving neurodegeneration in many diseases. Moreover, strategies to modulate autophagy have been shown to provide protection against neurodegeneration. Here we describe a novel and simple strategy to express an autophagy flux reporter in the nervous system of adult animals by the intraventricular delivery of adeno-associated viruses (AAV) into newborn mice. Using this approach we efficiently expressed a monomeric tandem mCherry-GFP-LC3 construct in neurons of the peripheral and central nervous system, allowing the measurement of autophagy activity in pharmacological and disease settings.
Similar content being viewed by others
Main
Macroautophagy, here referred to as autophagy, is the major catabolic pathway involved in the degradation of damaged or superfluous organelles, abnormal protein aggregates and other cytosolic components.1 An explosion of literature in the last few years highlights the contribution of autophagy to diverse physiological processes including metabolic control, lipid homeostasis, immunity and myopathies.2 Autophagy is initiated by the formation of autophagosomes, double membrane vesicles that engulf cytosolic components and fuse with endosomes to form hybrid organelles called amphisomes that later fuse with lysosomes where cargoes are degraded. The mechanisms underlying the regulation of different steps of autophagy are highly complex and dynamic and are reviewed in detail elsewhere.3, 4 In brief, two ubiquitin-like conjugation pathways control autophagy. One of them involves the covalent binding of Atg12 to Atg5, which occurs at the phagophore level, and dissociation after autophagosome consolidation. One of the key steps in the regulation of autophagy is the conjugation of microtubule-associated protein 1 light chain 3 (LC3) to phosphatidylethanolamine to form microtubule-associated protein 1 light chain 3 lipidated form (LC3-II). Lipidated LC3 binds to the expanding phagophore and remains associated with autophagosomes even after fusion with lysosomes. Then, LC3-II can be either delipidated and recycled or degraded by hydrolytic enzymes at the lysosome. Monitoring LC3-II conversion, LC3 distribution or its flux through the autophagy pathway are the gold standards for measuring autophagy activity and are widely used in the field.4
The generation of conditional knockout mice for essential autophagy regulatory components in the nervous system revealed a key role of the pathway in the basal maintenance of protein homeostasis in neurons.5, 6 Most prevalent neurodegenerative diseases, including Alzheimer’s disease, amyotrophic lateral sclerosis, Parkinson’s disease and Huntington’s disease are associated with the misfolding and aggregation of specific proteins.7 Many studies indicate that autophagy operates as an efficient mechanism for the degradation of aggregation-prone proteins linked to neurodegeneration and suggest that pharmacological activation of autophagy offers a promising therapeutic avenue.8 Accumulating evidence also suggests that autophagy impairment may underlie the etiology of several neurodegenerative diseases.8, 9
One of the main limitations in the field is the lack of reliable tools to monitor autophagy activity in the nervous system,4, 10 a challenging issue due to barriers for drug penetrance into the tissue through the blood–brain barrier (BBB), and the low response of this tissue to nutrient starvation. The accumulation of LC3-positive puncta by histological analysis or the measurement of LC3-II by western blot analysis in the brain tissue is commonly monitored as a measure of autophagy activation. However, it is well documented that interpretation of these analysis can be misleading because LC3 accumulation also results from autophagy impairment (i.e., decreased fusion to lysosomes or impaired cargo degradation).4 Moreover, a concomitant increase in both the rate of autophagosome formation and LC3-downstream degradation can show normal steady-state levels in LC3-II protein despite enhanced autophagy activity. Mice transgenic for GFP-LC3 have been generated and used to monitor autophagosome content in the nervous system,11, 12, 13, 14, 15, 16 which also has the same caveats for measuring autophagy activity described above.4 Here we develop a novel approach to deliver and express the dynamic autophagy flux reporter mCherry-GFP-LC317 in the nervous system using the intraventricular brain injection of adeno-associated vectors (AAV) in newborn mice. This method results in a wide distribution of the reporter in neurons of the central and peripheral nervous system with high efficiency,18 allowing the measurement of LC3 flux in vivo in single neurons. This integrative strategy offers a unique tool to assess autophagy activity in the context of the physiology of the nervous system, determine its contribution to diverse diseases and study how autophagy can be modified by therapeutic treatments.
Results
Widespread expression of GFP-mCherry-LC3 in the nervous system using AAV2 delivery system
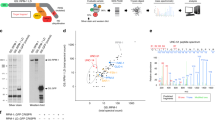
To develop an efficient strategy for expression of the autophagy flux reporter mCherry-GFP-LC3 in the nervous system of mice, we cloned this gene into an AAV expression vector (Figure 1a). This fluorescent reporter is designed to monitor LC3 flux through the autophagy pathway because the GFP signal is sensitive to the acidic and/or proteolytic conditions of the lysosome lumen, whereas mCherry fluorescence persists under these environments.4, 17 Therefore, the colocalization of both mCherry and GFP fluorescence signal, stains a vesicular compartment that has not fused with lysosomes (yellow puncta, i.e., phagophores or autophagosomes), whereas a mCherry vesicle without GFP fluorescence emission corresponds to an amphisome or autolysosome (red puncta, Figure 1a and the schematic pathway in Figure 1b). This reporter enables simultaneous estimation of both the induction of autophagy and LC3 flux without the use of lysosomal inhibitors. We selected AAV serotype 2 for efficient transduction of neurons in vivo and produced large-scale preparations using triple transfection of 293 cells using a rep/cap plasmid and pHelper. The activity of the sensor and the AAV were first tested in NSC34 motoneurons using western blot and spinning disk microscopy after autophagy stimulation with nutrient starvation by exposing cells to EBSS (Earle’s balanced salt solution) media (Figures 1c and d). Using a cocktail of lysosomal inhibitors, we were able to corroborate that this sensor can measure autophagy flux as detected by the accumulation of LC3-II by western blot analysis (Figure 1c), or the massive accumulation of autophagosomes (yellow dots) that did not reach lysosomes (Figure 1d).
Widespread delivery of AAV2/2_mCherry-GFP-LC3 through the nervous system. (a) Schematic representation of mCherry-GFP-LC3 AAV2/2 constructs. (b) Upon activation of autophagy, LC3 binds to phagophore to form the autophagosome (mCherry-GFP-LC3 generates yellow puncta). Then, autophagosomes fuse with lysosomes and amphisomes where the acidic environment quenches the green GFP fluorescence (compartment labeled in red). (c) NSC34 cells were transduced with AAV2/2_mCherry-GFP-LC3 for 72 h, followed by exposure to EBSS media to induce autophagy by severe nutrient starvation for indicated time points. mCherry-GFP-LC3 expression was monitored by western blot where mCherry-GFP-LC3-I and mCherry-GFP-LC3-II bands can be resolved. As control, cells were treated with a cocktail of lysosome inhibitors (bafilomycin, pepstatin and E64D; Lys. Inh.) for 6 h to monitor basal autophagy flux in these cells. Actin expression was monitored as loading control. (d) NSC34 cells described in (c) were fixed and visualized by spinning disk microscopy using GFP and TXRED filters. Scale bar: 10 μm. (e) Images of ICV injection of AAV into P1 pups. (f) Distribution of AAV2/2-GFP in the nervous system after ICV injection of newborn pups. Then animals were killed 4 weeks after injection for histological analysis. The following tissues were analyzed: (i) cortex, (ii) cerebellum, (iii) hippocampus, (iv) spinal cord ventral, (v) sciatic nerve and (v’) high magnification of indicated region of sciatic nerve to depict single axons
To obtain widespread gene delivery in the nervous system, we performed intracerebroventricular (ICV) injections of neonatal mice with 2 μl AAV2/2_GFP (titer 1.22 × 1012 DNase-resistant particles (DRP)/ml) (Figures 1e and f). This strategy has been shown to result in structure-specific distribution of the transgene with stable expression for at least 1 year, where the pattern of transduction is consistent with a global distribution of AAV2 through its circulation in the subarachnoid space via cerebral spinal fluid (CSF) flow.18, 19 At 4 weeks post-vector administration, animals were killed and the brain, spinal cord and sciatic nerves were analyzed by fluorescent microscopy. A widespread transduction of the cerebellum and other brain areas such as hippocampus and cortex was observed using this method (Figure 1f, see panels i–iii). In addition, the spinal cord was efficiently transduced with this strategy (Figure 1f, panel iv), showing a high neuronal tropism as assessed by the staining for the motoneuron marker choline acetyltransferase (ChAT) (Supplementary Figure 1a). Surprisingly, even peripheral neurons were transduced, as evidenced by the dissection of the sciatic nerve (Figure 1f, panel v and v́). On the basis of these results, we then applied ICV injection of AAV in neonatal animals to generate the widespread expression of mCherry-GFP-LC3 through the nervous system of mice and study the effect of different activators of autophagy (See Figure 2, next section).
Rapamycin and trehalose treatments increase LC3 flux through the autophagy pathway in the spinal cord. (a) Overview of the spinal cord of mice injected with AAV2/2_mCherry-GFP-LC3 after treatment or not with 2 g/kg of trehalose (plus 3% in drinking water, ad libitum) or 2 mg/kg of rapamycin, by 12 weeks. Images were taken with a × 5 super zoom objective in a confocal microscope, and two higher magnifications are shown at the right panels. Scale bars, right: 200 μm, middle: 40 μm, left: 10 μm. (b) Tissue from experiments presented in (a) were analyzed in parallel using spinning disk microscopy. Scale bar: 10 μm. (c) Quantification of LC3-positive dots presented in individual neurons were quantified using spinning disk microscopy and expressed as a percentage of cell with LC3 dots (minimum three dots per cell). Number of cells analyzed: control 40 (three animals), trehalose 112 (five animals) and rapamycin 86 (four animals). (d) Newborn animals were injected with AAV2/2_mCherry-GFP-LC3 and after 4 weeks treated or not with trehalose or rapamycin, by 12 weeks. Spinal cord tissue was then processed for histology and total GFP and mCherry fluorescence were measured in tissue sections of 140 × 140 μm in images obtained with a spinning disk microscope using × 40 objective to calculate LC3 fluxes by measuring fluorescence intensity of the tissue. (e) Using the same images, total GFP and mCherry fluorescence per cell area were measured to calculate LC3 flux. In panels c–e, data are presented as mean and S.E. Non-parametric t-test was used to compare statistical significance between drug treated and non-treated animals. **P<0.01; ***P<0.001
Rapamycin and trehalose treatments increase LC3 flux in neurons in vivo
We used confocal fluorescent and spinning disk microscopy to monitor the pattern of mCherry-GFP-LC3 distribution in mice transduced with AAV2 4 weeks following ICV injection. Visualization of spinal cord motoneurons in the ventral horn showed the basal presence of LC3-positive autophagosomes and autophagolysosomes, as depicted by fluorescent puncta in the cell soma (Figure 2a, upper right panel). We then treated mice with rapamycin, a classical inducer of mTOR-dependent autophagy, by administrating 2 mg/kg rapamycin through intraperitoneal (IP) injection at a frequency of three times per week, as previously used in the models of brain injury or prion disease.20, 21, 22 We confirmed that this rapamycin administration regimen decreases mTOR phosphorylation at ser 2481 in the brain cortex. This phosphorylation monitors mTOR autophosphorylation, and thus directly measures its catalytic activity (Supplementary Figure 2A). We also confirmed these findings by measuring the phosphorylation of the mTOR substrate p70S6K (phospho-threonine 371 (Supplementary Figure 2B). Similar results were obtained when the spinal cord tissue was analyzed (not shown). Afterward, we quantified three different parameters in the spinal cord motoneurons to measure autophagosome content and LC3 flux: (i) we measured the percentage of cells containing LC3-positive puncta to assess the number of cells responding to the stimuli; (ii) the ratio between the number of mCherry and yellow dots to monitor LC3 flux; and (iii) the total number of GFP and mCherry dots per cell to visualize total vesicular content. At basal levels, around 30% of AAV-transduced neurons contained LC3-positive vesicles, which was significantly increased after rapamycin administration reaching close to 70% of transduced cells (Figures 2b and c). Interestingly, rapamycin treatment slightly increased the overall content of LC3 dots (Figure 2c, right panel, see red bars), but it strongly stimulated the appearance of red LC3 puncta, indicating the occurrence of LC3 flux (Figure 2c, right panel). In addition, we were able to obtain similar results by quantifying total GFP and mCherry fluorescence in full spinal cord tissue sections or individual cells using an automatic measurement of the intensity using ImageJ software from the National Institutes of Health (http://rsbweb.nih.gov/ij/) (Figures 2d and e), indicating that this method can be used to quantify LC3 flux with different image analysis approaches.
mTOR-independent autophagy is perhaps a more interesting target for drug discovery,23, 24 as targeting this pathway may avoid the pleiotropic consequences of mTOR manipulation in a disease context.25 Trehalose is a disaccharide that enhances mTOR-independent autophagy in vitro and has been shown to have protective activity in many models of neurodegeneration.26, 27, 28, 29 However, because of technical limitations, no reports have shown the actual activation of autophagy flux in the nervous system by trehalose. Animals transduced with AAV2/2_mCherry-GFP-LC3 were IP injected with 2 g/kg trehalose three times a week, and this treatment was supplemented with ad libitum consumption of drinking water containing 3% trehalose. We recently used this treatment paradigm to induce neuroprotection on an ALS mouse model.30 Trehalose administration induced a dramatic effect in both autophagosome content (Figure 2) and LC3 flux in spinal cord neurons (Figures 2c–e). Remarkably, the pattern of distribution of red and green dots presented a different behavior from the results obtained after rapamycin administration (Figure 2c, right panel). These results indicate that our strategy to measure LC3 flux and phagosome content in vivo could discriminate specific effects of different stimulators of the autophagy pathway.
We then assessed the potential of our method to monitor LC3 flux in the brain and peripheral nerves. High expression of the AAV2/2_mCherry-GFP-LC3 construct was observed in the Purkinje cell layer of the cerebellum (Figures 3a and b), in addition to cortex, hippocampus and olfactory bulb with a high degree of reproducibility (Supplementary Figure 3, and not shown). We were able to detect autophagy enhancement and flux in the cerebellum of animals treated with rapamycin or trehalose where single cells were easily detected due to the high expression of the reporter gene (Figures 3a–c). Surprisingly, the effects of autophagy stimulation by drug treatment in the three parameters measured were clearly different in the same animals when LC3 puncta distribution was monitored in the cerebellum compared with the spinal cord (Figure 3c). In addition, the amplitude of the autophagy flux measured was significantly higher in Purkinje cells compared with spinal cord motoneurons (Figure 3c, middle panel). As the ectopic expression of LC3-GFP has been shown to generate protein aggregates in certain experimental systems that do not correspond to autophagy-related vesicles,4 we performed lysosomal staining using immunofluorescence with an anti-LAMP2 antibody and observed an evident colocalization with LC3/mCherry dots (Supplementary Figure 4).
Rapamycin and trehalose treatment increase LC3 flux in the cerebellum. (a) Visualization of the cerebellar crest of animals injected with AAV2/2_mCherry-GFP-LC3 treated or not with rapamycin or trehalose (left panel). Right panel: a magnified view of the Purkinje cell layers in the cerebellum the indicated area using a superzoom objective in a confocal microscope. (b) Tissue from experiments presented in (a) were analyzed in parallel using spinning disk microscopy. Scale bars, a: 100 μm (left) and 30 μm (right), b: 40 μm (left panel) and 10 μm (right panel). (c) LC3-positive vesicles and autophagy flux were quantified as described in Figure 2c. Mean and S.E. are presented. Number of cells analyzed: control 50 (three animals), trehalose 59 (five animals) and rapamycin 33 (four animals). Non-parametric t-test was used to compare statistical significance between drug treated and non-treated animals. **P<0.01; ***P<0.001
Autophagosome trafficking along axons was recently described.31 With this in mind, we monitored the distribution of mCherry-GFP-LC3 in the sciatic nerve of animals treated or not with the autophagy inducers. Single axons were easily identified in disaggregated explants of sciatic nerve, showing the presence of red and yellow LC3 puncta (Figure 4). In addition, drug treatments with trehalose or rapamycin also enhanced LC3 fluxes in these peripheral axons (Figure 4). These results demonstrate that the delivery of AAV2/2_mCherry-GFP-LC3 into the nervous system through ICV injection of newborn animals is a reliable and efficient method to monitor autophagy flux in neurons of adult mice.
Pharmacological activation of autophagy in the peripheral nerves. Sciatic nerves were obtained from animals transduced with AAV2/2_mCherry-GFP-LC3 and treated with trehalose or rapamycin as described in Figure 2. Sciatic nerve fibers were then dissociated and analyzed to visualize mCherry-GFP-LC3 distribution. White arrows indicate the presence of red dots along the fiber. Scale bar: 10 μm
Monitoring autophagy flux in mechanical injury models
In order to further validate our strategy for measuring autophagic flux in vivo, we monitored mCherry-GFP-LC3 in a pathological setting. Different reports suggest that autophagy is a relevant cellular response after spinal cord injury (SCI).32, 33, 34, 35 We performed SCI by a lateral hemisection of the spine in animals ICV-injected with AAV2/2_mCherry-GFP-LC3 as we recently described.36 Previous reports indicate that autophagy markers are upregulated 4 h after SCI, peaking at 3 days after surgury.32, 34 On the basis of this information, animals were killed 24 h post surgery and the spinal cord removed for histological analysis as illustrated in the scheme of Figure 5a. We observed an increase in sensor fluorescence in the injured region compared with control sham animals (Figure 5b). This local accumulation of the LC3 reporter was unexpected, and may possibly result from disrupted LC3 trafficking through the axon due to the mechanical injury. Analysis of single cells indicated a dramatic increase in the amount of LC3-positive autophagosomes and autophagolysosomes in this disease model (Figure 5b, right panels), confirming previous observations (examples in Sekiguchi et al.,33 Tanabe et al.37).
LC3 flux in the peripheral and CNS in models of mechanical injury. (a) Schematic diagram of the method and strategy employed to deliver AAV2/2_mCherry-GFP-LC3 by ICV followed by the challenge of mice with SCI or peripheral nerve crush. (b) Newborn animals were ICV-injected with AAV2/2_mCherry-GFP-LC3 particles and after 90 days the spinal cord was hemisected as described in Materials and Methods. After 24 h of recovery, mice were killed and the spinal cord slices were visualized with a confocal microscope. At the right panels, pictures taken with a superzoom objective in confocal microscope are shown to depict mCherry-GFP-LC3-positive dots in overlay images of the green and red channels: Three animals per groups were analyzed. Scale bars, right: 80 μm, middle: 20 μm, left: 8 μm (upper) and 6 μm (lower). (c) In parallel, 90 days after ICV injection of AAV2/2_mCherry-GFP-LC3 particles, mice were anesthetized and the sciatic nerve injury was performed by a nerve crush technique (see Materials and Methods). After 24 h of recovery, mice were killed and the sciatic nerve fibers were observed with a × 60 oil immersion objective in a spinning disk microscope. White arrows indicate mCherry-GFP-LC3 red dots along the fiber. Scale bar: 10 μm
We also tested our method to monitor autophagy in peripheral nerves. Autophagy has been suggested to be increased after sciatic nerve damage.38 It has been also proposed as a mechanism of axonal degeneration operating around 6 h post injury in the optic nerve.39 We performed sciatic nerve crush at the level of the sciatic nerve notch (Figure 5a), as we previously reported,40 and then visualized autophagy fluxes in single teased nerve fibers. The number of LC3 dots increased throughout the axons 24 h post injury (Figure 5c). Specifically in the damaged nerve, close to the injured area (∼3 mm distal), we observed a high enrichment of dots that were positive for mCherry only (Figure 5c), indicating that this method allows the visualization of the distribution of different autophagy-related structures throughout single axons of the sciatic nerve.
Measurement of single mCherry-GFP-LC3 vesicle trafficking
In order to apply our method to determine the dynamic of autophagic vesicles in axons, we transduced explants of dorsal root ganglia (DRG) with AAV2/2_mCherry-GFP-LC3, and after 24 h we treated this culture with rapamycin to perform a time-lapse analysis. Remarkably, a clear enhancement of autophagolysosome trafficking was observed through the axon after rapamycin administration (Figure 6a). Using a kinetogram analysis, we visualized the movement of LC3-positive vesicles (Figure 6b) and also calculated the average speed of single particles (Figure 6c), which ranged from 0.045 μm/s up to 0.9 μm/s in average. A significant enhancement of the average velocity was observed in the total population of LC3-possitive vesicles when DRG explants were treated with rapamycin (Figure 6c, right panel). Remarkably, the velocities obtained were in the same order of magnitude to the ones recently reported for autophagosomes in axons of DRGs obtained from LC3-GFP transgenic mice.31 As control for these experiments, we monitored by electron microscopy (EM) the presence of autophagy-related vesicles in axons, which were often associated with cytoskeleton fibers (Supplementary Figure 5). Thus, this method may offer also the opportunity to measure autophagy vesicle dynamics in ex vivo preparations of the brain and spinal cord tissue.
Autophagosome trafficking in axons from DRG explants. (a) Explants of DRG were transduced for 5 days with AAV2/2_mCherry-GFP-LC3 and analyzed by spinning disk microscopy. Then axons were recorded for 5 min and then stimulated with 400 nM rapamycin. Images of mCherry and GFP fluorescence are shown at indicated times after rapamycin administration. Merge images are also shown. White arrows indicate an example of a vesicle moving through the axon. (b) Kymograph of mCherry-GFP-LC3 motility in DRG axons before and after (indicated with a black arrow) rapamycin treatment. (c) Average speed of LC3-positive vesicle movement in axons of DRG explants described in a was determined for single particles. Left panel: vesicles speed distribution of a group of particles (events) before (white bars) and after (gray bars) rapamycin treatment. Right panel: average speed distribution of all vesicles described in the left panel. ***P<0.001
Discussion
In the study of neurodegenerative diseases, autophagy is emerging as a relevant target for drug discovery. In addition, due to the recent identification of specific defects in the autophagy pathway in diseases such as Alzheimer’s and Huntington’s disease,8 strategies to restore or bypass autophagy alterations represent novel therapeutic targets. Despite the fact that this research area has evolved very fast and new studies are published almost every week on this topic, most reports monitoring autophagy activity in the nervous system are based on the interpretation of static images of LC3 or p62 levels where the dynamics of the process have been difficult to assess. Many pharmacologic and genetic manipulations that enhance autophagy activity in cell culture models are usually correlated with effects of these manipulations in disease progression/protein aggregation in vivo to infer the role of autophagy in the nervous system. Here we describe a simple method to measure autophagy flux in the mouse nervous system by combining three different strategies: the use of high titers of AAV2/2, the dynamic fluorescent reporter mCherry-GFP-LC3 and the delivery of the particles using ICV injection in newborn animals. This approach led to widespread expression of the autophagy reporter in many different neuronal populations. mCherry-GFP-LC3-positive dots and autophagy flux were easily detected with different microscopy techniques (regular epifluorescence, confocal and spinning disk microscopy) after activation of the pathway with pharmacological inducers. As examples for possible applications of this method to disease conditions, we measured LC3 flux after SCI and peripheral nerve crush. In addition, this method was useful to determine the dynamic of autophagic vesicles along neuronal structures. We propose that our methodological principles may led to important applications in the neuroscience and biomedical field as it can be applied to any mouse model of diseases, it can be used to test the activity of drugs in vivo, or to define the effect of certain genetic manipulations on autophagy activity in neurons. The combinatorial method described here is simple and can be applied by any laboratory, as AAV production is now commercially available at a reasonable cost, and detailed and reliable protocols for ICV injections of viruses are available.18, 19
Overall autophagy activity in the nervous system has been indirectly inferred through a variety of techniques, including detection of autophagosomes by EM, the measure of static levels LC3-II by western blot, by the assessment of the distribution of autophagy-related proteins to vesicular structures or the decrease in the levels of autophagy substrates such as p62 or mutant Huntingtin.4 Because monitoring these markers can lead to misleading interpretations,4 there is a growing need for simple methods to measure autophagy flux in neurons in vivo. A recent study described a semi-quantitative method to measure LC3 fluxes in vivo in mice through the visualization of endogenous LC3 protein turnover in non-central nervous system tissues after the administration of leupeptin, a protease inhibitor.41 The nervous system was not analyzed in these experiments, probably because of the low permeability of this compound to the BBB after IP injection.42 Other inhibitors such as colchicine have also been employed to monitor LC3 turnover in the muscle.43 Colchicine requires 2 days of treatment, which is incompatible with the study of the autophagy process that occurs over minutes. In general, it is predicted that the use of general inhibitors of lysosomal proteases, microtubule dynamics or autophagosome-lysosome membrane fusion machinery will have devastating consequences to the cell’s function, representing a bulk perturbation of the system that has been measured. Our strategy allows monitoring different rates of autophagy activation in a subpopulation of cells and the identification of particular cell types undergoing autophagy activation or inhibition. Moreover, we were able to estimate the dynamic of LC3 vesicles in axons of DRG explants. Although our results showed that we are able to monitor LC3 flux in vivo after different stimulations, we cannot discard that a fraction of the vesicles/dots detected may correspond to LC3-mCherry-GFP aggregates of the fusion protein as this problem has been reported before in vitro after high overexpression.4 However, our measurement of vesicle trafficking and the colocalization with LAMP2 suggests that indeed the signals detected correspond to autophagic-related vesicles. As two-photon microscopy has been used to visualize neuronal process in vivo through the use of fluorescent probes,44 our method opens the possibility of measuring autophagosome dynamics in vivo in the context of the 3D structure of the brain in real time. We propose that the use of ICV injection of AAV2/2_mCherry-GFP-LC3 will represent an important tool in the advancement toward defining the contribution of autophagy in diverse physiological and pathological conditions affecting the nervous system.
Materials and Methods
Design and construction of AAV2/2 vectors
The mCherry-GFP-LC3 plasmid was kindly provided by Terje Johansen’s laboratory. The whole mCherry-GFP-LC3 expression cassette was excised from the plasmid and inserted into a shuttle pro-viral plasmid, containing both the AAV2/2 inverted terminal repeats (ITRs) and the 1.6-kb cytomegalovirus enhancer promoter. We produced large-scale preparations of AAV using triple transfection of 293T cells using a rep/cap plasmid and pHelper (Stratagene, La Jolla, CA, USA), followed by purification by column affinity chromatography.45 Finally, viral titers were quantified by real-time TaqMan PCR assay with primers that are specific for the bovine growth hormone polyadenylation (bgh-PolyA) sequence.
Cell culture and AAV2/2 transduction verification
We employed a motoneuron cell line (NSC34 cells) to verify the AAV2/2 mCherry-GFP-LC3 infectivity. Cells were transduced with a dilution 1 : 500 of AAV2/2 particles from 5.4 × 1012 DRP/ml stock preparations. After 72 h of transduction, cells were treated with EBSS for 1, 4 and 6 h (nutrient starvation). As a control, we treated the cells with a lysosome inhibitor cocktail (200 nM bafolimycin A1, and 10 μg/ml protease inhibitors pepstatin and E64D). Total cell extracts were prepared in RIPA buffer (20 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% DOC, 0.5% triton X-100) containing a protease inhibitor cocktail (Roche, Basel, Switzerland) by sonication. Protein concentration was determined by micro-BCA assay (Pierce, Rockford, IL, USA). The equivalent of 50 μg of total protein buffer containing protease inhibitors were analyzed by SDS-PAGE (12%) and western blot using an anti-LC3 antibody from Cell Signaling Technology, Inc (Danvers, MA, USA) in 1 : 1000 dilution (4 °C, O.N). Actin was used as a loading control 1 : 5000 (13E5, Cell Signaling).
ICV injections of AAV2/2
The mCherry-GFP-LC3 plasmid was described before17 and cloned into an AAV shuttle vector containing serotype 2 inverted terminal repeats. AAVs were produced using triple transfection of 293 cells using a rep/cap plasmid and pHelper (Stratagene), followed by purification by column affinity chromatography and, finally, we quantified of viral titers by real-time TaqMan PCR assay with primers that are specific for the bovine growth hormone polyadenylation (bgh-PolyA) sequence (Genzyme Corporation, Framingham, MA, USA). P0 to P2 pups received one injection of 2 μl AAV2/2_mCherry-GFP-LC3 into the cerebral lateral ventricles of the right hemisphere as described before18, 19 using a dose of 5.4 × 1012 DRP/ml. Briefly, P0 to P1 pups were cryo-anesthetized during 2 to 3 min and mounted over a LED-light to visualize the intersection of the coronal and the sagittal cranial sutures (bregma). The animal’s head was placed on a cold surface of Styrofoam covered with foil with a small saddle where the head was fixed to prevent movement (See Figure 1e). A fine-drawn capillary needle was inserted 1 mm lateral and 1 mm posterior to bregma, and then tunneled 1 mm deep to the skin edge (approximating) of ipsilateral ventricle. The animal was positioned head facing the researcher and injection was performed with the syringe directed ∼80° to the front and 10° to the center from the point of injection. A no toxic tracer (Fast Green, 0.04%) was added to the AAV2/2 content in the syringe to determine whether the ventricles were indeed injected and filled with the vectors. Only those pups in which the lateral ventricles were filled with viral solution were included in the study. P0 to P1 pups received one injection of 2 μl AAV2/2 mCherry-GFP-LC3 into the cerebral lateral ventricles of one of the hemispheres (regularly the right hemisphere). The AAV solution was injected at full strength with a total dose of 5.4 × 1012 DRP/ml. Following the injections, pups were put back to the mother until weaning. To prevent the rejection of the pups by the mother, pups were left carefully in shavings on the cage and allow to be found by the mother. All animals were used on a pure C57BL/6 genetic background and all treatments were approved by and carried out according to the guidelines of the University of Chile Institutional Animal Care and Use Committee.
Animal treatment and disease models
Mice were treated thrice by IP injections with 2 g/kg of trehalose (Sigma-Aldrich, St. Louis, MO, USA) or 2 mg/kg of rapamycin (LC Laboratories, Woburn, MA, USA) for indicated periods. In addition, trehalose was provided in the drinking water at 3% w/v at libitum (Tokyo Chemical Industry, Co., Ltd, America, Portland, OR, USA). Rapamycin was prepared in dimethyl sulfoxide (DMSO) (25 mg/ml) and stored at −20 °C until use. For animal injection, the stock solution was diluted immediately before injections with 0.5 ml aqueous solution containing 5% polyethylene glycol 400 (PEG400) and 5% Tween-80, and the solution volume was completed to with filtered phosphate-buffered saline (PBS) as described before in models of traumatic brain injury in mice.20 Trehalose for IP injections was prepared in 500 mg/ml in PBS solution, filtered, aliquot and frozen to −20 °C until use. In addition, trehalose was prepare at 3% w/v in the drinking water and was daily replace for a new fresh bottle. For the analysis of the efficacy of rapamycin treatment, western blot analysis was performed for the following antibodies and dilutions: phospho-mTOR (Ser2481), 1 : 1000 (catalog number 2971, Cell Signaling); mTOR 1 : 1000 (catalog number 2972, Cell Signaling); phospho-p70S6K (Thr389), 1 : 1000 (catalog number 9208, Cell Signaling); p70S6K 1 : 1000 (catalog number 9202, Cell Signaling).
Surgical procedure for SCI
Animals of 90 days after ICV injection were lateral hemisected as described before.36 In brief, all animals were anesthetized with an IP dose of avertin (tribromoethanol) (300 mg/kg). The dorsal zone of the spinal cord was incised along the midline and the spinal cord exposed at the T12 vertebra level where it was hemitransected on the right side using a vannas micro scissor (RS-5658, ROBOZ, Gaithersburg, MD, USA). Sham animals include the complete surgical procedure with no hemitransection of the spine. During recovery, mice were placed in a temperature-controlled chamber. Twenty-four hours post-surgery animals were perfused transcardially with 4% paraformaldehyde in 0.1 M PBS. A 5-mm region of the spinal cord containing the lesion site was removed and post-fixed for 3 h in 4% paraformaldehyde. The spinal tissue was subjected to a sucrose gradient (5, 10 and 30% sucrose in PBS), cryoprotected with optimal cutting temperature compound (Tissue-Tek, Alphen aan den Rijn, the Netherlands), and fast frozen using liquid nitrogen. Five millimeters of the spinal cord, around the injury area was cut longitudinally in 20 μm-thick sections using a cryostat microtome (Leica, Nussloch, Germany) and then visualized in a confocal microscope (Olympus FV1000, Olympus, Center Valley, PA, USA) with a × 5 to × 40 superzoom objective.
Surgical procedures for sciatic nerve crush
Ninety days after ICV injection with AAVs, C57BL/6 mice were anaesthetized with avertin (tribromoethanol) (300 mg/kg). Then, the right sciatic nerve was exposed at midthigh, freed from the surrounding tissues and then, was clamped in the notch region (crush injury) and then surgically seam. The nerve was crushed three times for 5 s with Dumont no. 5 forceps (Fine Science Tools INC, Foster City, CA, USA).40 Control animals were subjected to the same intervention with no crush. During recovery, mice were placed in a temperature-controlled chamber. After 24 h post-surgery, animals were euthanized and sciatic nerve removed for cryostat sections and dissociated fibers as described before.46 Pictures were taken in a spinning disk (Olympus XI 100) microscope with a × 60 oil immersion objective. Surgical interventions and animal care follows the Institutional Review Board’s Animal Care of Pontificia Universidad Católica de Chile.
Semiautomatic quantification of fluorescent dots in longitudinal slices from the spinal cord
Five millimeters length transversal sections of spinal cord from control mice or mice injected with AAVs were mounted into slides to analyze them in a spinning disk microscope. Images were acquired with a × 40 oil immersion objective using GFP and TXRED filters in a spinning disk microscope. We merged both channels for colocalization analysis (i.e., red and yellow puncta analysis). Manual counting of dots was performed on a double-blind manner in indicated number of animals and cells (see figure legends). In addition, for quantification of fluorescence intensity all the images of each experimental group were placed in a same image file and converted into a 16 bit image (black and white), in GFP and TXRED focal field images, respectively. Then, background signal was excluded determining a threshold over which fluorescent intensity was considered as positive in relation to negative controls, in the maximum slope of intensity using the ImageJ software. Then, we analyzed the integrated density of fluorescent signal in each channel, averaged and subjected to statistical test to determine the differences in LC3 flux between the experimental groups.
Histological and biochemical analysis
Four weeks after ICV, AAV2/2_GFP transduced mice were perfused transcardially with 4% paraformaldehyde in PBS. The spinal cord, sciatic nerve and the brain were removed and post-fixed for 3 h in 4% paraformaldehyde. The target tissues were subjected to a sucrose gradient (5, 10 and 30% sucrose in PBS), cryoprotected with optimal cutting temperature compound (Tissue-Tek) and fast frozen using liquid nitrogen. Tissues were cut using a cryostat microtome (Leica). Section were mounted with vectashield Hoechst nuclei stain and visualized in a confocal microscope (Olympus FV1000) using a × 5 superzoom objective. For single sciatic nerve fibers, tissue was removed and fixed for 1 h in 4% paraformaldehyde and dissected to obtain single fibers. The dissociation of the sciatic nerve was performed mechanically, with a tungsten needle. Then, fibers were mounted with 4′,6-diamidino-2-phenylindole (DAPI) nuclei stain and imaged in spinning disk (Olympus XI 100) microscope with a × 60 oil immersion objective. Spinal cord transversal section of animals injected with the AAV2/2_mCherry-GFP-LC3 and treated with rapamycin, trehalose and untreated were also imaged in a spinning disk (Olympus XI 100) microscope with a × 40 oil immersion objective. The tissue was longitudinally sectioned (5 μm thick slices) using a cryostat microtome (Leica). Sections were immunostained using antibodies anti-LAMP2 1 : 200 (ABL.93 hybridoma, Developmental Studies Hybridoma Bank, University of Iowa) and developed with a secondary antibody (alexa fluor 647 goat anti-mouse; Invitrogen, Carlsbad, CA, USA). Spectral confocal microscopy (Olympus FV1000 confocal microscope) was used to acquire images.
For analysis of phosphorylated mTOR, motor cortex was dissected and homogenized in RIPA buffer followed by Western analysis as described for NSC34 cells. The following antibodies and dilutions were used: anti-phospho-mTOR, mTOR, 1 : 2000, (Cell Signaling), and anti-Hsp90 (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
DRG culture
DRGs from the spinal cord of embryonic E16 Sprague Dawley rats were isolated in ice-cold Petri dishes containing L15 medium. Each DRG explants was plated in a well of a six-well plate and cultured for 4 days with the AAV2/2_mCherry-GFP-LC3 in Neurobasal medium (2% B27, 0.3% L-glutamine, 1% streptomycin/penicillin, 4 μM aphidicolin, 7.5 μg/ml of fluoro-2-deoxyuridine and 50 ng/ml of NGF) at 37 °C with 5% CO2. After 96 h, DRGs explants were treated with 400 nM rapamycin and visualized in a spinning disk microscope in a time-lapse mode. Images were analyzed using ImageJ software. Kymograph description and plugins using to calculate particle velocities can be downloaded from http://www.embl.de/eamnet/html/body_kymograph.html.
Electron microscopy
For electron microscope analysis, DRGs cultured in plastic coverslips were transduced with AAV2/2 mCherry-GFP-LC3. After 5 days, cells were fixed and processed using standard techniques for EM as described.47 Thin sections of Epon blocks were obtained and grids imaged with a Philips Tecnai 12 electron microscope (FEI, Hillsboro, OR, USA) operated at 80 kV and fitted with a digital camera as we described before.48
Statistical analysis
Statistical analyses used a two-tailed unpaired non-parametric Mann–Whitney test. P-values are specified in the figure legends. This was achieved to compare significance between untreated versus trehalose, and untreated versus rapamycin treatments.
Abbreviations
- AAV:
-
adeno-associated vectors
- LC3:
-
microtubule-associated protein 1 light chain 3
- LC3-II:
-
microtubule-associated protein 1 light chain 3 lipidated form
- DRP:
-
DNase-resistant particles
- ICV:
-
intracerebroventricular
- ChAT:
-
choline acetyltransferase
- EBSS:
-
Earle’s balanced salt solution
- IP:
-
intraperitoneal
- SCI:
-
spinal cord injury
- DRG:
-
dorsal root ganglia
- CNS:
-
central nervous system
- BBB:
-
blood–brain barrier
- DMSO:
-
dimethyl sulfoxide
- PBS:
-
phosphate-buffered saline solution
- DAPI:
-
4′,6-diamidino-2-phenylindole
References
Mizushima N, Levine B, Cuervo AM, Klionsky DJ . Autophagy fights disease through cellular self-digestion. Nature 2008; 451: 1069–1075.
Murrow L, Debnath J . Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol 2012; 8: 105–137.
Kroemer G, Marino G, Levine B . Autophagy and the integrated stress response. Mol Cell 2010; 40: 280–293.
Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544.
Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441: 880–884.
Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441: 885–889.
Matus S, Glimcher LH, Hetz C . Protein folding stress in neurodegenerative diseases: a glimpse into the ER. Curr Opin Cell Biol 2011; 23: 239–252.
Menzies FM, Moreau K, Rubinsztein DC . Protein misfolding disorders and macroautophagy. Curr Opin Cell Biol 2011; 23: 190–197.
Marino G, Madeo F, Kroemer G . Autophagy for tissue homeostasis and neuroprotection. Curr Opin Cell Biol 2011; 23: 198–206.
Mizushima N, Yoshimori T, Levine B . Methods in mammalian autophagy research. Cell 2010; 140: 313–326.
Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest 2008; 118: 2190–2199.
Katsumata K, Nishiyama J, Inoue T, Mizushima N, Takeda J, Yuzaki M . Dynein- and activity-dependent retrograde transport of autophagosomes in neuronal axons. Autophagy 2010; 6: 378–385.
Wang QJ, Ding Y, Kohtz DS, Mizushima N, Cristea IM, Rout MP et al. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci 2006; 26: 8057–8068.
Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y . In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 2004; 15: 1101–1111.
Tian F, Morimoto N, Liu W, Ohta Y, Deguchi K, Miyazaki K et al. In vivo optical imaging of motor neuron autophagy in a mouse model of amyotrophic lateral sclerosis. Autophagy 2011; 7: 985–992.
Mizushima N, Kuma A . Autophagosomes in GFP-LC3 Transgenic Mice. Methods Mol Biol 2008; 445: 119–124.
Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282: 24131–24145.
Passini MA, Wolfe JH . Widespread gene delivery and structure-specific patterns of expression in the brain after intraventricular injections of neonatal mice with an adeno-associated virus vector. J Virol 2001; 75: 12382–12392.
Glascock JJ, Osman EY, Coady TH, Rose FF, Shababi M, Lorson CL . Delivery of therapeutic agents through intracerebroventricular (ICV) and intravenous (IV) injection in mice. J Vis Experiments 2011; 56: e2968, doi:10.3791/2968.
Erlich S, Alexandrovich A, Shohami E, Pinkas-Kramarski R . Rapamycin is a neuroprotective treatment for traumatic brain injury. Neurobiol Dis 2007; 26: 86–93.
Cortes CJ, Qin K, Cook J, Solanki A, Mastrianni JA . Rapamycin delays disease onset and prevents PrP plaque deposition in a mouse model of Gerstmann-Straussler-Scheinker disease. J Neurosci 2012; 32: 12396–12405.
Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nature Genet 2004; 36: 585–595.
Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC . Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ 2009; 16: 46–56.
Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat Chem Biol 2008; 4: 295–305.
Sengupta S, Peterson TR, Sabatini DM . Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell 2010; 40: 310–322.
Schaeffer V, Lavenir I, Ozcelik S, Tolnay M, Winkler DT, Goedert M . Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain 2012; 135: 2169–2177.
Rodriguez-Navarro JA, Rodriguez L, Casarejos MJ, Solano RM, Gomez A, Perucho J et al. Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neurobiol Dis 2010; 39: 423–438.
Davies JE, Sarkar S, Rubinsztein DC . Trehalose reduces aggregate formation and delays pathology in a transgenic mouse model of oculopharyngeal muscular dystrophy. Hum Mol Genet 2006; 15: 23–31.
Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC . Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem 2007; 282: 5641–5652.
Castillo K, Nassif M, Valenzuela V, Rojas F, Matus S, Mercado G et al. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy 2013; 9: 1–13.
Maday S, Wallace KE, Holzbaur EL . Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol 2012; 196: 407–417.
Kanno H, Ozawa H, Sekiguchi A, Itoi E . Spinal cord injury induces upregulation of Beclin 1 and promotes autophagic cell death. Neurobiol Dis 2009; 33: 143–148.
Sekiguchi A, Kanno H, Ozawa H, Yamaya S, Itoi E . Rapamycin promotes autophagy and reduces neural tissue damage and locomotor impairment after spinal cord injury in mice. J Neurotrauma 2012; 29: 946–956.
Kanno H, Ozawa H, Sekiguchi A, Yamaya S, Itoi E . Induction of autophagy and autophagic cell death in damaged neural tissue after acute spinal cord injury in mice. Spine (Phila Pa 1976) 2011; 36: E1427–E1434.
Chen HC, Fong TH, Lee AW, Chiu WT . Autophagy is activated in injured neurons and inhibited by methylprednisolone after experimental spinal cord injury. Spine (Phila Pa 1976) 2012; 37: 470–475.
Valenzuela V, Collyer E, Armentano D, Parsons GB, Court FA, Hetz C . Activation of the unfolded protein response enhances motor recovery after spinal cord injury. Cell Death Dis 2012; 3: e272.
Tanabe F, Yone K, Kawabata N, Sakakima H, Matsuda F, Ishidou Y et al. Accumulation of p62 in degenerated spinal cord under chronic mechanical compression: functional analysis of p62 and autophagy in hypoxic neuronal cells. Autophagy 2011; 7: 1462–1471.
Penas C, Font-Nieves M, Fores J, Petegnief V, Planas A, Navarro X et al. Autophagy, and BiP level decrease are early key events in retrograde degeneration of motoneurons. Cell Death Differ 2011; 18: 1617–1627.
Knoferle J, Koch JC, Ostendorf T, Michel U, Planchamp V, Vutova P et al. Mechanisms of acute axonal degeneration in the optic nerve in vivo. Proc Natl Acad Sci USA 2010; 107: 6064–6069.
Barrientos SA, Martinez NW, Yoo S, Jara JS, Zamorano S, Hetz C et al. Axonal degeneration is mediated by the mitochondrial permeability transition pore. J Neurosci 2011; 31: 966–978.
Haspel J, Shaik RS, Ifedigbo E, Nakahira K, Dolinay T, Englert JA et al. Characterization of macroautophagic flux in vivo using a leupeptin-based assay. Autophagy 2011; 7: 629–642.
Osanai T, Nagai Y . Suppression of experimental allergic encephalomyelitis (EAE) with liposome-encapsulated protease inhibitor: therapy through the blood-brain barrier. Neurochem Res 1984; 9: 1407–1416.
Ju JS, Varadhachary AS, Miller SE, Weihl CC . Quantitation of "autophagic flux" in mature skeletal muscle. Autophagy 2010; 6: 929–935.
Nimmerjahn A, Kirchhoff F, Helmchen F . Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 2005; 308: 1314–1318.
Anderson R, Macdonald I, Corbett T, Whiteway A, Prentice HG . A method for the preparation of highly purified adeno-associated virus using affinity column chromatography, protease digestion and solvent extraction. J Virol Methods 2000; 85: 23–34.
Court FA, Zambroni D, Pavoni E, Colombelli C, Baragli C, Figlia G et al. MMP2-9 cleavage of dystroglycan alters the size and molecular composition of Schwann cell domains. J Neurosci 2011; 31: 12208–12217.
Court FA, Midha R, Cisterna BA, Grochmal J, Shakhbazau A, Hendriks WT et al. Morphological evidence for a transport of ribosomes from Schwann cells to regenerating axons. Glia 2011; 59: 1529–1539.
Castillo K, Rojas-Rivera D, Lisbona F, Caballero B, Nassif M, Court FA et al. BAX inhibitor-1 regulates autophagy by controlling the IRE1alpha branch of the unfolded protein response. Embo J 2011; 30: 4465–4478.
Acknowledgements
We are indebted to Omar Ramirez and Carmen Gloria Lemus for assistance in the use of the confocal superzoom microscope. This work was funded primary by FONDEF D11I1007, Millennium Institute No. P09-015-F, Ring Initiative ACT1109, CONICYT-EEUU No. USA 2013-0003, FONDECYT no. 1100176 (CH); FONDECYT Postdoctoral Fellowship no. 3100112 and Ring Initiative ACT1104 (KC). Current address: Centro Interdisciplinario de Neurociencia de Valparaíso (CINV), Facultad de Ciencias, Universidad de Valparaíso; FONDECYT 11121524 (SM). We also received funding from the Muscular Dystrophy Association, the Michael J Fox Foundation for Parkinson’s Research, ALS Therapy Alliance and Alzheimer Association (CH), CONICYT and FEBA Foundation PhD fellowships (MN), CONICYT graduate fellowship (VV), FONDECYT no. 1110987 and Millennium Nucleus no. P07-011-F (FAC). BLS is supported by the Swiss National Science Foundation, grant 31003A_135696. The Biomedical Neuroscience Institute and The Centro Interdisciplinario de Neurociencia de Valparaíso are Millennium Institutes supported by the Millennium Scientific Initiative of the Ministerio de Economía, Fomento y Turismo.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Verkhratsky
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Castillo, K., Valenzuela, V., Matus, S. et al. Measurement of autophagy flux in the nervous system in vivo. Cell Death Dis 4, e917 (2013). https://doi.org/10.1038/cddis.2013.421
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.421
Keywords
This article is cited by
-
Reduction of DHHC5-mediated beclin 1 S-palmitoylation underlies autophagy decline in aging
Nature Structural & Molecular Biology (2024)
-
CTB-targeted protocells enhance ability of lanthionine ketenamine analogs to induce autophagy in motor neuron-like cells
Scientific Reports (2023)
-
Lysosomal dysfunction in Schwann cells is involved in bortezomib-induced peripheral neurotoxicity
Archives of Toxicology (2023)
-
A distinct astrocyte subtype in the aging mouse brain characterized by impaired protein homeostasis
Nature Aging (2022)
-
Molecular imaging of the kinetics of hyperactivated ERK1/2-mediated autophagy during acquirement of chemoresistance
Cell Death & Disease (2021)