Abstract
In response to intense stress, the tumor protein p53 (p53) tumor suppressor rapidly mounts a direct mitochondrial death program that precedes transcription-mediated apoptosis. By eliminating severely damaged cells, this pathway contributes to tumor suppression as well as to cancer cell killing induced by both genotoxic drugs and non-genotoxic p53-reactivating molecules. Here we have explored the role had in this pathway by the prolyl-isomerase Pin1 (peptidylprolyl cis/trans isomerase, NIMA-interacting 1), a crucial transducer of p53’s phosphorylation into conformational changes unleashing its pro-apoptotic activity. We show that Pin1 promotes stress-induced localization of p53 to mitochondria both in vitro and in vivo. In particular, we demonstrate that upon stress-induced phosphorylation of p53 on Ser46 by homeodomain interacting protein kinase 2, Pin1 stimulates its mitochondrial trafficking signal, that is, monoubiquitination. This pathway is induced also by the p53-activating molecule RITA, and we demonstrate the strong requirement of Pin1 for the induction of mitochondrial apoptosis by this compound. These findings have significant implications for treatment of p53-expressing tumors and for prospective use of p53-activating compounds in clinics.
Similar content being viewed by others
Main
Among the biological activities of tumor protein p53 (p53), induction of apoptosis has a prominent role in tumor suppression by eliminating incipient cancer cells and in mediating cytotoxic effects that in part determine the success of conventional cancer treatments. p53-dependent apoptosis has a pleiotropic nature; in addition to affecting the balance between pro- and anti-apoptotic factors at the transcriptional level, p53 also directly modulates the functions of some of these factors at the mitochondria, thus triggering successive waves of mitochondrial outer membrane permeabilization (MOMP) in response to acute and sustained stress conditions.1, 2 A common early event in p53-mediated apoptosis induced by cytotoxic and hypoxic conditions is the rapid (within 30 min) translocation of cytoplasmic p53 to mitochondria,3 where it interacts with members of the Bcl family of mitochondrial permeability regulators, including Bak, Bcl2 and Bcl-XL, to either inhibit or activate them (see ref. 1) for a comprehensive review). This causes MOMP and apoptosis that precedes target gene activation by p53.4 It has been clearly shown that transcription-independent apoptotic activity of p53 is important for tumor suppression.5, 6, 7 However, the exact mechanism responsible for stress-induced mitochondrial accumulation and activation of p53 is not yet fully understood.
We previously demonstrated that upon severe stress stimuli, the activity of the prolyl-isomerase Pin1 (peptidylprolyl cis/trans isomerase, NIMA-interacting 1) is crucial for enabling p53 to take the apoptotic route of stress response.8, 9, 10 Pin1 catalyzes cis/trans isomerization of proline bonds preceded by phosphorylated serine or threonine residues (pSer/Thr-Pro), thereby altering structure and functions of its substrates in response to specific cues.11, 12 Upon genotoxic insults, Pin1 binds multiple sites on p53, promoting its dissociation both from HDM2 with consequent accumulation in stressed cells,8, 13 and from the apoptosis inhibitor iASPP through isomerization of the phospho-Ser46-Pro47 motif, thus unleashing p53’s full apoptotic potential.9, 10 However, the impact of Pin1 isomerization in controlling p53 function by altering its sub-cellular trafficking has never been addressed.
With this work, we shed light on the mechanisms promoting p53’s direct mitochondrial death program by demonstrating a central role for the prolyl-isomerase Pin1 in regulating this p53 apoptotic route in response to several stress signals.
Results
Pin1 potentiates the transcription-independent apoptotic activity of p53
To verify whether Pin1 could modulate the direct apoptotic activity of p53 at mitochondria, we employed a nuclear import-deficient p53 construct (p53NLS, Figure 1a) shown to efficiently localize at mitochondria and induce apoptosis when overexpressed in p53-null cancer cells.14 At first, we decided to investigate the impact of p53 and Pin1 on the mitochondrial calcium (Ca2+) response after agonist stimulation as a highly sensitive readout of mitochondrial state. Indeed, mitochondrial alterations cause defects in Ca2+ uptake by the organelle.15 To this end, we employed a mitochondria-targeted chimera of the Ca2+-sensitive photoprotein aequorin.16
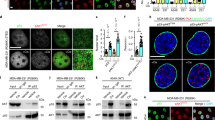
Pin1 potentiates p53-mediated transcription-independent apoptosis. (a) Scheme of p53 expression constructs indicating Pin1-binding sites (phospho-Ser/Thr-Pro). TA, transactivation domain; DBD, DNA binding domain; NLS, nuclear localization signal. The nuclear import-deficient p53 mutant p53NLS has mutations in all three C-terminal NLS motifs. p53-M bears Ser/Thr to Ala substitutions at Pin1 binding sites Ser33, Ser46, Thr81 and Ser315. p53NLS-M bears Ser/Thr to Ala substitutions at Ser33, Ser46 and Thr81. (b) Mitochondrial Ca2+ analysis in H1299 cells overexpressing p53NLS alone or co-transfected with Pin1. H1299 cells were co-transfected with an aequorin chimera targeted to the mitochondrial matrix and with either the plasmid of interest or an empty vector (control). Thirty-six hours after transfection, measurement of aequorin luminescence was carried out and calibrated into [Ca2+] values, as described in the Materials and Methods section. Where indicated, the cells, perfused with KRB, were challenged with 100 μM ATP, added to the same buffer. Peak values 1.58±0.03 μM for control (solid line) n=14; 1.18±0.09 μM for p53 NLS (dotted line) n=13, P<0.01; p53NLS plus Pin1 (dashed line) 0.93±0.04 μM, n=9, P<0.01. These and the following traces are representative of at least 10 experiments, that gave similar results. (c) p53NLS was overexpressed in p53-null H1299 cells either alone or along with Pin1-HA. The percentage of cells undergoing apoptosis upon treatment with etoposide 50 μM for 24 h was estimated by AnnexinV/propidium-iodide staining and FACS analysis. The graph shows mean results and S.D. of three independent experiments. (d) Mitochondrial Ca2+ response was performed as above, comparing H1299 cells overexpressing p53NLS with p53NLS M proteins. Peak values: control and p53NLS are as in b. p53 NLS-M 1.57±0.05 μM, n=12. (e) The ability of p53NLS and p53NLS-M proteins to induce transcription-independent apoptosis was compared by AnnexinV/propidium-iodide staining and FACS analysis upon overexpression in p53-null H1299 cells and treatment with etoposide 50 μM for 24 h. The graph shows mean results and S.D. of three independent experiments. (f) The effect of Pin1 overexpression on release of cytochrome c from mitochondria to the cytoplasm was detected by WB after subcellular fractionation of H1299 cells transfected with the indicated constructs and treated with etoposide 50 μM for 24 h
We investigated the Ca2+ response to ATP, which acts on Gq-coupled plasmamembrane receptors and causes the production of inositol 1,4,5 trisphosphate, thus releasing Ca2+ from the ER to mitochondria. We observed that the overexpression of p53NLS alone caused a 31% decrease in amplitude of the Ca2+ spike evoked by agonist stimulation as compared with control cells, as a consequence of mitochondrial damage. More interestingly, co-overexpression of Pin1 further potentiated mitochondrial Ca2+ alterations induced by p53NLS, thus amplifying the mitochondrial damage (Figure 1b). Similar results were obtained in HCT116 p53−/− cells (Supplementary Figure 1G).
We then tested the effect of Pin1 on the ability of p53NLS to exert transcription-independent apoptosis. Annexin V-FACS analysis and poly (ADP-ribose) polymerase (PARP) cleavage assay highlighted that co-expression of Pin1 caused a significant increase of apoptosis induced by p53NLS upon etoposide treatment (Figure 1c and Supplementary Figure 1H). Conversely, p53NLS-dependent apoptosis was reduced by twofold following short interfering RNA (RNAi)-mediated knockdown of Pin1 expression (Supplementary Figure 1A). Activation of the mitochondrial apoptotic pathway was demonstrated by the release of cytochrome c from mitochondria to the cytoplasm, and notably this was higher in cells overexpressing Pin1 (Figure 1f).
To confirm the role of Pin1 in transcription-independent apoptosis induced by endogenous p53, we treated HCT116 p53+/+ cells with the RNA polymerase II inhibitor α-amanitin, which was previously shown to completely block p53-dependent transcription (Supplementary Figure 1B), yet induce apoptosis via the p53 mitochondrial program.3, 17 RNAi-mediated knockdown of Pin1 expression resulted in significant reduction of transcription-independent apoptosis induced by etoposide treatment (Supplementary Figure 1C).
We next asked whether direct modification of p53 by Pin1 is required for enhancing its transcription-independent apoptotic activity, similar to what occurs with nuclear p53 during induction of apoptotic target genes.9 Cytotoxic stress conditions in fact strongly promote the interaction of cytoplasmic p53NLS with Pin1 (Supplementary Figures 1D and 3A). We generated a p53NLS construct with Ser/Thr-to-Ala substitutions disrupting the major Pin1-binding sites (phospho-Ser/Thr-Pro)8, 9 at residues 33, 46 and 81 (p53NLS-M, Figure 1a), thus severely reducing the binding to Pin1 (Supplementary Figure 1E). Notably, in contrast to p53NLS, overexpression of p53NLS-M had little or no effect on the mitochondrial Ca2+ uptake of H1299 cells (Figure 1d). Moreover, the ability of p53NLS-M to induce apoptosis upon treatment with etoposide was significantly reduced as compared with p53NLS (Figure 1e). In further support, as compared with wild-type full-length p53, p53-M (Figure 1a) was almost unable of inducing transcription-independent apoptosis when overexpressed in the presence of α-amanitin (Supplementary Figure 1F). Taken together, these data suggest that modification of p53 by Pin1 is an essential step for optimal execution of transcription-independent p53-mediated apoptosis at mitochondria.
Pin1 is required for efficient mitochondrial localization of p53
A key step during induction of transcription-independent apoptosis is the relocalization of cytoplasmic p53 to mitochondria in response to apoptotic stimuli.3, 18 Thus, we investigated whether modification by Pin1 affects the mitochondrial localization of p53 induced by DNA damage. Mitochondria were isolated from HCT116 p53+/+ cells after treatment with cytotoxic doses of chemotherapeutic drugs for 6 h. As compared with untreated cells, both doxorubicin and etoposide treatments caused accumulation of p53 at mitochondria, in agreement with published data,18 whereas the mitochondrial fraction of Pin1 remained unchanged (Supplementary Figure 2A). Notably, mitochondrial accumulation of p53 in drug-treated cells was strongly increased by Pin1 overexpression and reduced by RNAi-mediated knockdown of Pin1 (Figure 2a, and Supplementary Figure 2A and B). Importantly, the overexpression of siRNA-resistant wild-type Pin1 rescued p53’s mitochondrial translocation, whereas a catalytic inactive Pin1 mutant (S67E) failed to exert this effect (Figure 2a). This highlights the importance of Pin1 isomerase activity in inducing p53’s proapoptotic function. In further support, in vitro translated p53 was able to bind its mitochondrial partner BclXL only after isomerization by catalytically active Pin1 (Supplementary Figure 2C).
Pin1 stimulates mitochondrial localization of p53. (a) The effect of Pin1 depletion and overexpression on mitochondrial accumulation of endogenous p53 protein was analyzed by WB on both mitochondrial fractions and total lysates of HCT116 p53+/+ cells. Cells were transduced with retroviral vector pMSCV expressing either Pin1-HA, siRNA-resistant (SR) Pin1-HA, siRNA-resistant Pin1-HA-S67E (catalytically inactive) or empty vector, and then transfected with Pin1-specific RNAi or control RNAi. Forty-eight hours after transfection, cells were treated with doxorubicin (Dox) 1 μM for 6 h and processed for subcellular fractionation. (b) Rapid mitochondrial localization of p53 in cardiac muscle in vivo was compared in wt (Pin1+/+) and Pin1-KO (Pin1−/−) mice treated IP with 20 mg/kg of doxorubicin for 3 h. Western blots show p53 content in total lysate (TOT) of heart tissue and in mitochondrial fraction (MITO). Mitochondrial purity from nuclear contamination was verified by Lamin B WB. (c) The ability of p53-WT and p53-M proteins (Figure 1a) to localize at mitochondria was compared as in Figure 2a upon transfection in HCT116 p53−/− cells and treatment with doxorubicin (Dox) 1 μM for 6 h. (d) The effect of Pin1 on the balance between mono- and polyubiquitinated forms of p53 was analyzed in H1299 cells transfected with constructs expressing p53, HA-ubiquitin and Pin1 in the indicated combinations, and then treated with doxorubicin 1 μM for 6 h and with the proteasome inhibitor MG-132 50 μM for 4 h before IP and WB analysis
Direct mitochondrial activity of p53 has been implicated in radiation and drug sensitivity in vivo; genotoxic treatments induce mitochondrial accumulation of p53 in radio- and chemosensitive mouse organs, leading to rapid apoptosis;4 moreover, inhibition of mitochondrial p53 activity protects mice from effects of lethal γ-radiation.19 Thus, we sought to determine the impact of Pin1 on the p53-dependent mitochondrial response in vivo. To this end, we treated wild-type and Pin1-KO mice with a cardiotoxic dose of doxorubicin, and analyzed early mitochondrial accumulation of p53 in cardiac muscle, a tissue particularly rich in mitochondria and a major site of doxorubicin-related toxicity. Strikingly, although p53 accumulated to high levels at mitochondria of treated wild-type mice, this response was absent in tissues lacking Pin1 (Figure 2b), suggesting that Pin1 is essential for mitochondrial localization and mitochondrial functions of p53 in vivo.
Likewise, impaired mitochondrial accumulation upon doxorubicin and defective transcription-independent apoptosis was also observed in HCT116 p53−/− cells transfected with the p53-M mutant lacking the major Pin1-binding sites (Figure 2c and Supplementary Figure 1F), confirming that direct modification by Pin1 alters p53’s ability to associate with mitochondria. Importantly, after lethal irradiation of wild-type and Pin1-KO mice, bone marrow cells derived from Pin1-KO animals were resistant to apoptosis as compared with wild-type counterparts (Supplementary Figure 2D). This difference was however not observed for thymocytes, thus suggesting a potential tissue-specific proapoptotic activity of Pin1 in vivo.
One important post-translational modification regulating mitochondrial translocation of p53 is monoubiquitination.3 Interestingly, Pin1 was found to affect p53’s mono- versus polyubiquitination ratio.20 Importantly, consistent with this notion, upon overexpression of Pin1, we observed a marked decrease of the polyubiquitinated p53 pool in H1299 cells treated with doxorubicin 1 μM for 6 h (Figure 2d, analyzed in the presence of the proteasome inhibitor MG132 for optimal visualization). Conversely, overexpression of Pin1 favored the increase of monoubiquitinated p53 species (Figure 2d). This observation may explain the mechanism by which Pin1 drives the mitochondrial localization of p53.
Pin1 induces mitochondrial function of p53 in a Ser46-dependent fashion
To gain insights into the mechanism by which modification by Pin1 may increase p53 monoubiquitination, we first sought to determine which Pin1-target site(s) on p53 is responsible for activating p53-mediated mitochondrial apoptosis. Ser46 phosphorylation is determinant for transcriptional apoptotic activity of p5321, 22 induced by a Pin1-dependent mechanism.9, 10 We generated a cytosolic p53 construct mimicking the absence of Ser46 phosphorylation (p53NLS-S46A). As compared with its wild-type counterpart, p53NLS-S46A displayed greatly reduced binding to Pin1 (Supplementary Figure 3A), suggesting that phospho-Ser46-Pro47 represents a major Pin1-binding site on cytosolic p53. p53NLS-S46A displayed reduced mitochondrial localization in doxorubicin-treated cells (Figure 3a and Supplementary Figure 3B) and was consequently impaired for mitochondrial activities, that is, alteration of the mitochondrial Ca2+ uptake (Figure 3b) and induction of transcription-independent apoptosis (Figure 3c). Importantly, the weak apoptotic activity of p53NLS-S46A could not be potentiated by overexpression of Pin1 (Figure 3c), indicating that Ser46 phosphorylation is indeed the determinant for inducing mitochondrial apoptosis by a Pin1-dependent mechanism. Consistent with this notion, Pin1 was able to increase monoubiquitination of wild type, but not of S46A-mutated p53 (Supplementary Figure 2E).
Pin1 induces the mitochondrial function of p53 in a Ser46-dependent fashion. (a) The ability of p53-WT, p53-S46A and p53-M proteins to localize at mitochondria was compared by WB on the mitochondrial fraction upon transfection in HCT116 p53−/− cells and treatment with etoposide 50 μM for 6 h. Left panels: a representative experiment is shown. Right panel: quantification of relative mitochondrial localization of different p53 proteins was calculated as described in Methods section. The graph shows the mean results and S.D. of three independent experiments. (b) Mitochondrial Ca2+ response was performed as above, comparing H1299 cells overexpressing p53NLS with p53NLS S46A proteins. Peak values 1.57±0.03 μM for control, 1.51±0.06 μM for p53NLS S46A, n=10 each, versus 1.20±0.04 μM for p53NLS, P<0.01. (c) The ability of p53NLS and p53NLS-S46A proteins to induce transcription-independent apoptosis upon treatment with etoposide 50 μM for 24 h, and the relative effect of overexpression of Pin1-HA was compared by AnnexinV/PI staining and FACS analysis upon transfection of the indicated combinations of plasmids in H1299 cells. The graph shows the mean results and S.D. of three independent experiments
HIPK2 and Pin1 cooperate to induce direct apoptotic activity of p53 at mitochondria
The above results suggest that p53’s apoptotic activity at mitochondria is triggered by upstream stimuli inducing Ser46 phosphorylation. In fact, a 6-h treatment with doxorubicin, which is sufficient to induce mitochondrial accumulation of p53 (Supplementary Figure 2A), also triggers its robust phosphorylation on this Pin1-binding site (Supplementary Figure 4A). In these conditions, we observed stabilization of the proapoptotic homeodomain-interacting protein kinase-2 (HIPK2; Supplementary Figure 4B), which phosphorylates p53 at Ser46.23, 24 We thus hypothesized that HIPK2 could act as an important mediator in activating both the nuclear and mitochondrial apoptotic pathways of p53.
To directly address the question whether HIPK2 might promote p53 localization to mitochondria, we knocked down HIPK2 expression by RNAi in HCT116 p53+/+ cells and consequently observed a decrease of stress-induced mitochondrial accumulation of p53 (Figure 4a and Supplementary Figure 4C). Conversely, overexpression of wild-type HIPK2 kinase, but not of the catalytic inactive mutant HIPK2-K221R, potentiated doxorubicin-induced accumulation of p53 at mitochondria (Figure 4b and Supplementary Figure 4C). Notably, Pin1 appears to be essential for this effect, as HIPK2 failed to trigger mitochondrial translocation of p53 upon Pin1 silencing (Figure 4c). Confirming this notion, transcription-independent apoptosis triggered by forced expression of HIPK2 in the presence of α-amanitin also required Pin1 (Figure 4d). In summary, these data suggest that upon genotoxic stress, rapid phosphorylation on Ser46 mediated by HIPK2 drives early activation of p53. Pin1 is then required to further modify Ser46-phosphorylated p53, thus enabling its translocation to mitochondria and triggering direct apoptosis.
HIPK2 and Pin1 cooperate to induce p53-dependent direct apoptosis. (a) HIPK2 promotes mitochondrial localization of p53. Accumulation of endogenous p53 within the mitochondrial fraction of HCT116 p53+/+ cells was analyzed by WB upon RNAi-mediated knockdown of HIPK2 and treatment with doxorubicin 1 μM for 6 h. C: control RNAi. (b) Mitochondrial localization of endogenous p53 upon overexpression of either wild-type HIPK2 or the catalytically inactive mutant HIPK2K221R in HCT116 p53+/+ cells and treatment with 1 μM doxorubicin for 6 h was analyzed as in a. (c) Pin1 is required for the ability of HIPK2 to promote mitochondrial localization of p53. The effect of wild-type HIPK2 overexpression on the mitochondrial localization of endogenous p53 was evaluated as in a in HCT116 p53+/+ cells treated with doxorubicin 1 μM for 6 h upon RNAi-mediated knockdown of Pin1 expression. (d) To estimate the roles of HIPK2 and Pin1 in inducing transcription-independent apoptosis, HCT116 p53+/+ cells transfected with the indicated combinations of GFP-HIPK2 expression vector and Pin1-specific RNAi were treated with α-amanitin 10 μg/ml. Apoptosis of GFP-expressing cells was then evaluated by TUNEL assays. The graph shows the mean results and S.D. of three independent experiments
Pin1 is essential for transcription-independent apoptosis induced by RITA in transformed cells
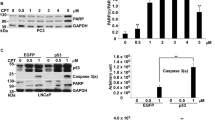
The small molecule RITA (reactivation of p53 and induction of tumor cell apoptosis) has been described as being able to activate wild-type p53’s apoptotic function with consequent antitumor activity. Mechanistically, RITA disrupts the p53-HDM2 complex by binding to p5325 and induces HIPK2 stabilization with consequent phosphorylation of p53 on Ser46.26 On these premises, we investigated the ability of RITA to induce p53’s direct mitochondrial activity. We analyzed cell viability upon administration of RITA in combination with α-amanitin to HCT116 p53+/+ and HCT116 p53−/− cells. RITA induced HIPK2 stabilization and consequent phosphorylation of p53 on Ser46 (Supplementary Figure 5A) and caused p53-dependent cytotoxicity (Figure 5a, and Supplementary Figure 5B and C) through both transcription-dependent and -independent pathways (Figures 5a and c). Indeed, a clear accumulation of endogenous p53 was observed in the mitochondrial fraction of RITA-treated HCT116 p53+/+ cells (Figure 5b). Induction of p53-Ser46 phosphorylation (Rinaldo et al26 and Supplementary Figure 5A) was required for the pro-apoptotic effect of RITA, given that the p53-S46A mutant failed to efficiently localize at mitochondria in response to RITA treatment (Supplementary Figure 5D). Importantly, reducing Pin1 expression with RNAi strongly inhibited RITA-induced mitochondrial p53 accumulation (Figure 5b) and transcription-independent apoptosis (Figure 5c). The observed requirement of Pin1 for RITA-dependent cytotoxicity may offer a possible explanation for RITA’s selective apoptotic effect in tumor cells, as opposed to its inefficiency in normal cells.25, 27 Indeed, Pin1 is frequently overexpressed in many common cancers, including mammary, prostate, colorectal and hepatic (28 and references therein). To test the role of Pin1 in determining the outcome of RITA treatment in transformed versus non-transformed cells, we employed MCF10A normal mammary epithelial cells where either HRASV12 or empty vector were stably inserted. Consistent with the notion that Pin1 expression is enhanced by HRAS,29, 30 MCF10A-HRASV12 cells exhibited elevated Pin1 levels as compared with parental cells (Figure 5d). Treatment with RITA led to significant induction of apoptosis (as estimated by PARP cleavage) only in RAS-transformed cells (Figure 5d), and importantly, this response could be efficiently inhibited by reducing Pin1 expression by RNAi (Figure 5e). Consistently, subcellular fractionation highlighted that RITA treatment induced mitochondrial accumulation of p53 in RAS-transformed cells (Figure 5f) but not in control MCF10A cells (Supplementary Figure 5E). This effect was significantly reduced by depleting Pin1 by RNAi (Figure 5f). Of note, overexpression of Pin1 in parental MCF10A cells was sufficient to shift the response to RITA treatment towards apoptosis (Supplementary Figure 5F). Taken together, these data strongly suggest that Pin1 expression levels are an important determinant of cancer cell sensitivity to RITA-induced cytotoxicity by favoring direct mitochondrial apoptosis.
Pin1 expression is required for RITA-induced transcription-independent apoptosis. (a) HCT116 p53+/+ cells were treated with RITA 1 μM alone or in combination with α-amanitin 10 μg/ml or left untreated. Cell viability was analyzed after 24 h. The graph shows the mean results and S.D. of three independent experiments. (b) The effect of Pin1 depletion by RNAi on mitochondrial localization of endogenous p53 protein upon treatment of HCT116 p53+/+ cells with RITA 1 μM for 24 h was analyzed by WB of mitochondrial fractions and total cell lysates. C: control RNAi. (c) HCT116 p53+/+ cells were transfected with Pin1-specific or control (C) siRNA before treatment with RITA 1 μM and α-amanitin 10 μg/ml for 24 h. The percentage of cells with sub-G1 DNA content (apoptotic cells) was estimated by PI staining and FACS analysis. The graph shows the mean results and S.D. of three independent experiments. (d) MCF10A cells stably transfected with either HRASV12 or empty vector (pLPC) were treated with RITA 1 μM for 24 h. Induction of apoptosis was then monitored by analyzing PARP cleavage by WB. The expression levels of HIPK2, p53, RAS and Pin1 were also compared by WB, using actin levels as loading control. (e) The effect of Pin1 depletion by RNAi on apoptosis of MCF10A-HRASV12 transformed cells upon treatment with RITA 1 μM for 24 h was analyzed by monitoring PARP cleavage by WB. (f) The effect of Pin1 depletion by RNAi on mitochondrial localization of endogenous p53 protein upon treatment of MCF10A-HRASV12 transformed cells with RITA 1 μM for 24 h was analyzed by WB of both mitochondrial fractions and total cell lysates
Discussion
Recent years have brought a significant advancement in understanding the contribution of various partners and co-factors in determining numerous aspects of the p53 pathway.31, 32 Among these, the prolyl-isomerase Pin1, a phospho-specific transducer of post-translational modifications for numerous key signaling molecules, surely is one of the most interesting.12 Although many evidences established the essential role of Pin1 in the transcription-dependent functions on p53 in the nucleus,8, 9, 33 to our knowledge no data have supported an involvement of Pin1 in other key aspects of p53’s growth-suppressive activities, such as its mitochondrial death program.
In this work, we demonstrate the strong requirement of this isomerase in the very early phases of p53-dependent apoptosis by controlling its mitochondrial accumulation and activation. The transcription-independent death program of p53 may be regarded as an attractive pathway to be exploited for therapeutic intervention. Therefore, in addition to provide insights into the yet poorly understood mechanisms of p53-dependent mitochondrial apoptosis, our results also carry implications for the effectiveness of anticancer treatments.
A role for Pin1 at the mitochondria level has been shown in controlling the activity of apoptosis regulators such as Bcl2,34 BIMEL35 and p66Shc16 in response to various extra- and intracellular stimuli. Our results demonstrating that, through isomerization of Ser46-phosphorylated p53, Pin1 promotes its mitochondrial trafficking signal (i.e., monoubiquitination) and translocation to these organelles further extends the cellular functions of the Pin1 isomerase. It is well known that by promoting p53 prolyl-isomerization, Pin1 lowers its affinity for HDM2 (8), and that low cellular levels of HDM2 favor p53 monoubiquitination and mitochondrial translocation.3, 36 Moreover, it has been shown that Pin1 inhibition results in decreased p53 monoubiquitination in favor of polyubiquitination.20 Together, these and our evidences raise the possibility that by priming Pin1-dependent conformational changes of p53, phosphorylation at Ser46 on p53 represents a key event in promoting its monoubiquitination and underlines the pivotal role of Pin1 in p53 mitochondrial relocalization and function. In this regard, we have shown that phosphorylation of p53 on Ser46 by the pro-apoptotic kinase HIPK2 is both necessary and sufficient for subsequent Pin1-mediated engagement of p53 to mitochondria where, facilitated by resident HDMX, it may then interact with Bcl2 as proposed by Mancini et al.37 Pin1 has been reported to localize in nucleus, cytoplasm and mitochondria, and it may act both in the cytoplasm, by triggering mitochondrial translocation of p53, and directly on the mitochondria, by increasing the retention of p53 in this compartment.
Our findings might also explain the reported impact of the p53 codon 72 Pro/Arg polymorphism on direct mitochondrial activity of p53. The Arg72 isoform displays higher mitochondrial translocation,38 and indeed, we have found this variant to be more phosphorylated on Ser46 and to bind Pin1 better than the Pro72 counterpart.9 Our work thus confirms a widespread crucial role for Ser46 phosphorylation in governing p53’s apoptotic activity at all levels, by triggering the rapid mitochondrial response to acute cytotoxic stress, as well as enabling the subsequent transcriptional activation of apoptotic target genes upon prolonged death-inducing stimuli. However, it is conceivable that in addition to HIPK2, other cytoplasmic pro-apoptotic kinases triggering p53 Ser46 phosphorylation (e.g., PKC-δ) could similarly induce, through Pin1, the localization of p53 at mitochondria.10, 39
Of note, we have recently found that HIPK2 engages p53 to induce the death of neurons expressing mutant forms of the protein Huntingtin.10 In light of the results presented here, a possible contribution of p53 transcription-independent apoptosis to neuronal death in Huntington’s disease could be envisaged.
The finding that a common set of stress-responsive factors (i.e., HIPK2 and Pin1) governs both the transcription-independent and -dependent arms of p53’s apoptotic program emphasizes the cooperative nature of these phases in fully executing the cellular response to stress stimuli. It can be envisioned that following a first wave of direct mitochondrial permeabilization induced by the HIPK2/Pin1/p53 pathway, prolonged signaling along the same axis reaches the threshold for activating p53 target genes, thus amplifying the response through a full apoptotic program (Figure 6).
Model for regulation of both transcription-dependent and -independent apoptotic activity of p53 by Pin1. Treatment with either chemotherapeutic drugs or RITA leads to stabilization of the apoptotic kinase HIPK2 and consequent phosphorylation of p53 on Ser46. Subsequent prolyl-isomerization by Pin1 then leads to decreased polyubiquitination of p53 in favor of its monoubiquitination, with consequent relocalization of cytosolic p53 to mitochondria and induction of MOMP and direct apoptosis. On the other hand, Pin1-mediated isomerization of the Ser46-Pro47 site unlocks p53 from the apoptosis inhibitor iASPP, leading to induction of apoptotic target genes and establishment of a full apoptotic response
This said, not all cell types are equally prone to undergo transcription-independent apoptosis. In vivo, radiation-sensitive tissues respond to DNA-damaging treatments with mitochondrial localization of p53 and rapid apoptosis, whereas radio-resistant tissues fail to undergo these processes.4 Moreover, Pin1 appears to be required for p53-dependent apoptosis in some tissues, whereas not in others. It would be interesting to explore the basis of this tissue specificity, possibly related to different kinases phosphorylating p53.
Besides its activity in a normal context, we have demonstrated that Pin1 is a crucial determinant for the sensitivity of oncogene-transformed cells to the p53- and HIPK2-activating drug RITA. We have observed that in tumor-derived cell lines, RITA induces mitochondrial accumulation of p53 and transcription-independent apoptosis. This is in line with studies where inhibition of p53’s mitochondrial activity by pifithrin-μ19 was able to reduce RITA’s cytotoxic effects in cancer cell lines.40, 41 On the basis of our observations that this process requires Pin1, one may anticipate that tumors displaying high levels of Pin1 might be more sensitive to RITA-dependent cytotoxicity, thus increasing the chances of selectively targeting cancer cells and minimizing undesired side effects. Of note, also the p53-activating molecule Nutlin has been shown to mainly induce p53-mediated transcription-independent apoptosis.7 It would be of interest to evaluate the role of Pin1 in this pathway, given that Ser46 phosphorylation appears to be a common requirement for the cytotoxic action of both Nutlin and RITA in cancer cells.42
As a consequence of its enzymatic nature, Pin1’s function is strongly influenced by the cellular context and it may therefore support the function of either tumor suppressors or oncogenic substrates. We have recently reported that in cancer cells, Pin1 acts to fully unleash the oncogenic potential of mutant p53.30, 43 Remarkably, RITA has been shown to confer transcription-independent apoptotic activity at least to some p53 mutant proteins,40 and also it might therefore prove effective in treatment of tumors bearing mutant p53 and high levels of Pin1.
Finally, besides its death-inducing potential, p53 localization at mitochondria may have broader tumor suppressive roles, related to inhibition of autophagy44 and tumor cell survival, as well as to metabolism control.45 Exploring the implications of the mitochondrial connection between p53 and Pin1 has therefore the potential to disclose further complexity.
Materials and Methods
Cell lines and treatments
Human colon carcinoma HCT116 p53+/+ and p53−/− and p53-null H1299 non-small cell lung carcinoma cell lines were maintained in DMEM or RPMI (H1299) with 10% FCS. MCF10A and MCF10A-HRasV12 were maintained in DMEM/F12 Ham’s (Sigma-Aldrich, St. Louis, MO, USA) 1 : 1, supplemented with 5% horse serum (Gibco, Life Technologies, Darmstadt, Germany), insulin (10 μg/ml; Sigma-Aldrich), hydrocortisone (0.5 μg/ml) and EGF (20 ng ml–1; Peprotech, Rocky Hill, NJ, USA). α-amanitin (Sigma-Aldrich), MG132 (Sigma-Aldrich), doxorubicin (Ebewe Pharma, Holzkirchen, Germany), etoposide (Sigma-Aldrich), RITA (Cayman, Ann Arbor, MI, USA), Ubiquitin Aldehyde (Enzo Life Biosciences, Vinci, Italy) were used at the concentrations indicated in the text.
Transfection and retroviral transduction
Lipofectamine 2000 (Invitrogen, Life Technologies, Darmstadt, Germany) was used for DNA transfection; for RNAi, double-stranded RNA oligonucleotides (10 pmol/cm2) were transfected using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer’s instructions.
Oligonucleotide sequences are reported below:

Retroviruses were made by calcium phosphate transfection of 293-GP packaging cells with the appropriate plasmids in combination with pMD2ENV coding for envelope proteins, and harvested 48 h later. Infected cells were selected with puromycin or blasticidin 2 mg/ml.
Expression plasmids
pcDNA3-Pin1, pcDNA3-Pin1-HA, pGEX-Pin1WW, pcDNA3-p53 WT, pcDNA3-p53-M and pcDNA3p53-S46A have been previously described.8, 10 pcDNA3-p53NLS was generated by subcloning from pCMV BamNeo-p53NLS.3 pcDNA3-p53NLS-M mutant was generated by PCR-directed mutagenesis. pLPC-Pin1-HA was generated by subcloning. HA-Ubiquitin expression vector was provided by PP DiFiore (IFOM, Milan, Italy). pCMV-Flag-HIPK2 WT, pEGFP-HIPK2 WT and pCMV-Flag-K221R was provided by S. Soddu (IRE Rome, Italy). pLPC-HRASV12 was provided by M. Serrano (CNIO, Madrid, Spain). pMSCV-HA Pin1SR and pMSCV-HA Pin1SR-S67E were generated introducing silent mutations in the region targeted by Pin1 siRNA by site-directed mutagenesis of pcDNA3-HAPin1 or pcDNA3-HAPin1S67E, and then subcloning in pMSCV vector.
Apoptosis and viability assays
Apoptosis was determined by cytofluorimetry using the Annexin V-FICT apoptosis detection kit (Sigma-Aldrich). For propidium iodide (PI) staining, cells were permeabilized with 0.1% NP40 in PBS and 2 μg/ml RNaseA, and treated with 10 μg/ml PI before analysis by flow cytometry. At least 1.5 × 104 cells were analyzed in each acquisition and the percentage of cells with DNA content <2n was calculated. TUNEL assays were performed with TMR Red in situ Cell Death Detection Kit (Roche, Nutley, NJ, USA) following manufacturer’s instructions. For viability assays, cells placed in 96-well plates (3000 cells per well) and treated with indicated compounds were incubated with WST-1 reagent (Roche) and further analyzed according to the manufacturer’s instructions.
Mitochondrial Ca2+ response: aequorin measurements
The cells were grown on coverslips and transfected with an aequorin chimera targeted to the mitochondrial matrix (mtAEQ). After 1–2 h of incubation with 5 μM coelenterazine, the coverslips were transferred to the perfusion chamber.
All aequorin measurements were carried out in KRB (Krebs-Ringer modified buffer: 125 mM NaCl, 5 mM KCl, 1 mM Na3PO4, 1 mM MgSO4, 5.5 mM glucose, 20 mM HEPES, pH 7.4, 37 °C) supplemented with 1 mM CaCl2. Agonists were added to the same medium. The experiments were terminated by lysing the cells with 100 μM digitonin in a hypotonic Ca2+-rich solution (10 mM CaCl2 in H2O), thus discharging the remaining aequorin pool. The light signal was collected and calibrated into [Ca2+] values by an algorithm based on the Ca2+ response curve of aequorin at physiological conditions of pH, [Mg2+] and ionic strength, as previously described.16 Representative traces are shown in the figures, whereas the full data set is included in the legends.
In vitro binding, immunoprecipitation and western blotting
Western blot (WB) analysis, immunoprecipitations and GST pull-down assays were performed as previously described.8 For ubiquitination assays, cells were lysed in 2% SDS, 150 mM NaCl, 10 mM Tris-HCl, pH 8.0, 1 mM PMSF, 5 mM NaF, 1 mM Na3VO4, 0,5% (v/v) sodium deoxycholate with protease inhibitor cocktail (Sigma-Aldrich) and Ubiquitin Aldeyde 50 ng/ml. Cell lysates were diluted in IP buffer: 10 mM Tris-HCl, pH8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton. Anti-p53 393FL (Santa Cruz Biotechnology, Santa Cruz, CA, USA) antibody was covalently bound to protein G Sepharose (Amersham Biosciences, GE Healthcare, Munich, Germany) using 5 mg/ml dimethylpimelimidate (Pierce Biosciences, Thermo Fisher Scientific, Bonn, Germany). Other antibodies were anti-p53 DO-1 (Santa Cruz Biotechnology), monoclonal anti-Pin1 (G8, Santa Cruz Biotechnology) and polyclonal anti-Pin1,8 anti-PARP p85 (Promega, Mannheim, Germany), anti-Porin VDAC (31HL, Calbiochem, Merck, Darmstadt, Germany), anti-lamin B1 (ABI6048), anti-PCNA (FL-261, Santa Cruz Biotechnology), polyclonal anti-actin (C11, Sigma-Aldrich), anti-p53pS46 (Cell Signaling), anti-HA 12CA5, anti-cleaved-caspase 3 (Cell Signaling, Danvers, MA, USA). Anti-HIPK2 antibody was kindly provided by T Hoffman (DKFZ, Heidelberg, Germany). For WB, equal protein amounts of mitochondrial and crude cell lysates were loaded. In cases where WBs were normalized for equal p53 loading, a first quantitation WB was run.
Mitochondria purification
Mitochondria were isolated by differential centrifugation, using Mitochondria Fractionation Kit (Pierce Biosciences). Mitochondrial purity was verified by WB for absence of nuclear (PCNA or Lamin B) contamination; equal mitochondrial loading was verified by WB for the mitochondrial resident porin VDAC. Quantification of relative mitochondrial localization was performed by densitometry analysis of WBs using the ImageJ analysis tool. Absolute intensity (AI) for each experimental p53 band and corresponding control band was obtained, and relative intensity was calculated by normalizing the experimental AI to the corresponding control AI. Mitochondrial p53 band was normalized both for mitochondrial extraction (mitochondrial VDAC band) and for transfection efficiency.
Immunofluorescence staining was performed as previously described.32 Briefly, cells were fixed in 4% paraformaldehyde for 20 min, washed in PBS, and blocked in PBS and BSA 10% for 30 min. Antigen recognition was done by incubating with anti-p53 DO1 as primary antibody for 1 h at 37 °C and with anti-mouse Alexa Fluor 594 as secondary antibody for 30 min a 37 °C.
Colocalization analysis
Colocalization between mt-GFP and p53 NLS (nuclear localization signal) proteins was calculated using ImageJ (rsb.info.nih.gov/ij) with the plug-in: Manders’ Coefficients. Colocalization was expressed as the Pearson’s correlation coefficient. For each condition, 20 single-cell images were collected by using confocal microscope Zeiss LSM 510 (Zeiss, Göttingen, Germany) and processed for colocalization analysis.
In vitro transcription and translation and in vitro isomerization
In vitro transcription and translation of MBP and MBP-p53 proteins was performed by using the TNT transcription-translation system (Promega) following the manufacturer’s instructions. In vitro isomerization was performed by incubating purified MBP-p53 proteins with 200 ng of either GST–Pin1WT, GST–WW or GST in the following buffer: 50 mM HEPES, pH7.5, 100 mM NaCl, 1 mM MgCl2, 1 mM dithiothreitol, supplemented with phosphatase inhibitors, for 20 min at room temperature.
Mice strains and mitochondria purification from mouse tissues
Pin1 KO mice in C57Bl background46 were provided by A. Means (Duke University, Durham, NC, USA). Genotyping was performed by PCR on tail DNA as described.46 Mice were injected IP with a single dose of either doxorubicin at 20 mg/kg or the same volume of saline as a control. Mice were killed after 3 h, and heart tissues were subsequently collected and processed for subcellular fractionation as described.47
For irradiation, mice were subjected to whole-body γIR with 10 Gy. Mice were killed 3 h after the treatment, and organs were immediately harvested and processed for WB.
Abbreviations
- MOMP:
-
mitochondrial outer membrane permeabilization
- Ca2+:
-
calcium
- NLS:
-
nuclear localization signal
- RNAi:
-
short interfering RNA
- RITA:
-
reactivation of p53 and induction of tumor cell apoptosis
- Pin1:
-
peptidylprolyl cis/trans isomerase, NIMA-interacting 1
- p53:
-
tumor protein p53
- HIPK2:
-
homeodomain interacting protein kinase 2
- PARP:
-
poly (ADP-ribose) polymerase
References
Vaseva AV, Moll UM . The mitochondrial p53 pathway. Biochim Biophys Acta 2009; 1787: 414–420.
Green DR, Kroemer G . Cytoplasmic functions of the tumour suppressor p53. Nature 2009; 458: 1127–1130.
Marchenko ND, Wolff S, Erster S, Becker K, Moll UM . Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J 2007; 26: 923–934.
Erster S, Mihara M, Kim RH, Petrenko O, Moll UM . In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol 2004; 24: 6728–6741.
Talos F, Petrenko O, Mena P, Moll UM . Mitochondrially targeted p53 has tumor suppressor activities in vivo. Cancer Res 2005; 65: 9971–9981.
Palacios G, Crawford HC, Vaseva A, Moll UM . Mitochondrially targeted wild-type p53 induces apoptosis in a solid human tumor xenograft model. Cell Cycle 2008; 7: 2584–2590.
Vaseva AV, Marchenko ND, Moll UM . The transcription-independent mitochondrial p53 program is a major contributor to nutlin-induced apoptosis in tumor cells. Cell Cycle 2009; 8: 1711–1719.
Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S et al. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 2002; 419: 853–857.
Mantovani F, Tocco F, Girardini J, Smith P, Gasco M, Lu X et al. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat Struct Mol Biol 2007; 14: 912–920.
Grison A, Mantovani F, Comel A, Agostoni E, Gustincich S, Persichetti F et al. Ser46 phosphorylation and prolyl-isomerase Pin1-mediated isomerization of p53 are key events in p53-dependent apoptosis induced by mutant huntingtin. Proc Natl Acad Sci USA 2011; 108: 17979–17984.
Yeh ES, Means AR . PIN1 the cell cycle and cancer. Nat Rev Cancer 2007; 7: 381–388.
Lu KP, Zhou XZ . The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol 2007; 8: 904–916.
Zheng H, You H, Zhou XZ, Murray SA, Uchida T, Wulf G et al. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 2002; 419: 849–853.
Marchenko ND, Moll UM . The role of ubiquitination in the direct mitochondrial death program of p53. Cell Cycle 2007; 6: 1718–1723.
Pinton P, Giorgi C, Siviero R, Zecchini E, Rizzuto R . Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene 2008; 27: 6407–6418.
Pinton P, Rimessi A, Romagnoli A, Prandini A, Rizzuto R . Biosensors for the detection of calcium and pH. Methods Cell Biol 2007; 80: 297–325.
Arima Y, Nitta M, Kuninaka S, Zhang D, Fujiwara T, Taya Y et al. Transcriptional blockade induces p53-dependent apoptosis associated with translocation of p53 to mitochondria. J Biol Chem 2005; 280: 19166–19176.
Marchenko ND, Zaika A, Moll UM . Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem 2000; 275: 16202–16212.
Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I et al. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol 2006; 2: 474–479.
Siepe D, Jentsch S . Prolyl isomerase Pin1 acts as a switch to control the degree of substrate ubiquitylation. Nat Cell Biol 2009; 11: 967–972.
Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 2000; 102: 849–862.
Mayo LD, Seo YR, Jackson MW, Smith ML, Rivera Guzman J, Korgaonkar CK et al. Phosphorylation of human p53 at serine 46 determines promoter selection and whether apoptosis is attenuated or amplified. J Biol Chem 2005; 280: 25953–25959.
D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 2002; 4: 11–19.
Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W et al. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol 2002; 4: 1–10.
Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med 2004; 10: 1321–1328.
Rinaldo C, Prodosmo A, Siepi F, Moncada A, Sacchi A, Selivanova G et al. HIPK2 regulation by MDM2 determines tumor cell response to the p53-reactivating drugs nutlin-3 and RITA. Cancer Res 2009; 69: 6241–6248.
Grinkevich VV, Nikulenkov F, Shi Y, Enge M, Bao W, Maljukova A et al. Ablation of key oncogenic pathways by RITA-reactivated p53 is required for efficient apoptosis. Cancer Cell 2009; 15: 441–453.
Finn G, Lu KP . Phosphorylation-specific prolyl isomerase Pin1 as a new diagnostic and therapeutic target for cancer. Curr Cancer Drug Targets 2008; 8: 223–229.
Ryo A, Liou YC, Wulf G, Nakamura M, Lee SW, Lu KP . PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol Cell Biol 2002; 22: 5281–5295.
Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell 2011; 20: 79–91.
Collavin L, Lunardi A, Del Sal G . p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ 2010; 17: 901–911.
Drost J, Mantovani F, Tocco F, Elkon R, Comel A, Holstege H et al. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat Cell Biol 2010; 12: 380–389.
Berger M, Stahl N, Del Sal G, Haupt Y . Mutations in proline 82 of p53 impair its activation by Pin1 and Chk2 in response to DNA damage. Mol Cell Biol 2005; 25: 5380–5388.
Basu A, Das M, Qanungo S, Fan XJ, DuBois G, Haldar S . Proteasomal degradation of human peptidyl prolyl isomerase pin1-pointing phospho Bcl2 toward dephosphorylation. Neoplasia 2002; 4: 218–227.
Becker EB, Bonni A . Pin1 mediates neural-specific activation of the mitochondrial apoptotic machinery. Neuron 2006; 49: 655–662.
Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W . Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 2003; 302: 1972–1975.
Mancini F, Di Conza G, Pellegrino M, Rinaldo C, Prodosmo A, Giglio S et al. MDM4 (MDMX) localizes at the mitochondria and facilitates the p53-mediated intrinsic-apoptotic pathway. EMBO J 2009; 28: 1926–1939.
Dumont P, Leu JI, Della Pietra AC, George DL, Murphy M . The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 2003; 33: 357–365.
Coutinho I, Pereira C, Pereira G, Gonçalves J, Côrte-Real M, Saraiva L . Distinct regulation of p53-mediated apoptosis by protein kinase Cα, δ, ɛ and ζ: evidence in yeast for transcription-dependent and -independent p53 apoptotic mechanisms. Exp Cell Res 2011; 317: 1147–1158.
Zhao CY, Grinkevich VV, Nikulenkov F, Bao W, Selivanova G . Rescue of the apoptotic-inducing function of mutant p53 by small molecule RITA. Cell Cycle 2010; 9: 1847–1855.
Kazemi A, Safa M, Shahbazi A . RITA enhances chemosensivity of pre-B ALL cells to doxorubicin by inducing p53-dependent apoptosis. Hematology 2011; 16: 225–231.
Ma T, Yamada S, Ichwan SJ, Ohtani K, Otsu M, Ikeda MA . Inability of p53-reactivating compounds Nutlin-3 and RITA to overcome p53 resistance in tumor cells deficient for p53Ser46 phosphorylation. Biochem Biophys Res Commun 2012; 417: 931–937.
Napoli M, Girardini JE, Piazza S, Del Sal G . Wiring the oncogenic circuitry: Pin1 unleashes mutant p53. Oncotarget 2011; 2: 654–656.
Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 2008; 10: 676–687.
Vousden KH, Ryan KM . p53 and metabolism. Nat Rev Cancer 2009; 9: 691–700.
Atchison FW, Capel B, Means AR . Pin1 regulates the timing of mammalian primordial germ cell proliferation. Development 2003; 130: 3579–3586.
Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P . Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc 2009; 4: 1582–1590.
Acknowledgements
We thank A Grison, A Bisso, A Rustighi and G Pastore for technical support. R Vidimari and A Beorchia for help in mice irradiation. We thank A Testa for editing the manuscript. This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC), Regione Friuli-Venezia Giulia, Italian University and Research Ministerium (Cofin FIRB) to GDS and by grants from AIRC and Telethon to PP.
Author contributions
FM and GDS designed and supervised the research and analyzed data; GS, MM, NR and CG performed the research; PP supervised CG and analyzed data; UM contributed new reagents and analyzed data; FM and GDS wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Sorrentino, G., Mioni, M., Giorgi, C. et al. The prolyl-isomerase Pin1 activates the mitochondrial death program of p53. Cell Death Differ 20, 198–208 (2013). https://doi.org/10.1038/cdd.2012.112
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2012.112
Keywords
This article is cited by
-
Depletion of mitochondrial methionine adenosyltransferase α1 triggers mitochondrial dysfunction in alcohol-associated liver disease
Nature Communications (2022)
-
Cancer and Alzheimer’s disease inverse relationship: an age-associated diverging derailment of shared pathways
Molecular Psychiatry (2021)
-
Ser46 phosphorylation of p53 is an essential event in prolyl-isomerase Pin1-mediated p53-independent apoptosis in response to heat stress
Cell Death & Disease (2019)
-
miR-663 sustains NSCLC by inhibiting mitochondrial outer membrane permeabilization (MOMP) through PUMA/BBC3 and BTG2
Cell Death & Disease (2018)
-
Prolyl isomerase Pin1: a promoter of cancer and a target for therapy
Cell Death & Disease (2018)