Abstract
Accumulating data indicate that the ubiquitin–proteasome system controls apoptosis by regulating the level and the function of key regulatory proteins. In this study, we identified Trim17, a member of the TRIM/RBCC protein family, as one of the critical E3 ubiquitin ligases involved in the control of neuronal apoptosis upstream of mitochondria. We show that expression of Trim17 is increased both at the mRNA and protein level in several in vitro models of transcription-dependent neuronal apoptosis. Expression of Trim17 is controlled by the PI3K/Akt/GSK3 pathway in cerebellar granule neurons (CGN). Moreover, the Trim17 protein is expressed in vivo, in apoptotic neurons that naturally die during post-natal cerebellar development. Overexpression of active Trim17 in primary CGN was sufficient to induce the intrinsic pathway of apoptosis in survival conditions. This pro-apoptotic effect was abolished in Bax−/− neurons and depended on the E3 activity of Trim17 conferred by its RING domain. Furthermore, knock-down of endogenous Trim17 and overexpression of dominant-negative mutants of Trim17 blocked trophic factor withdrawal-induced apoptosis both in CGN and in sympathetic neurons. Collectively, our data are the first to assign a cellular function to Trim17 by showing that its E3 activity is both necessary and sufficient for the initiation of neuronal apoptosis.
Similar content being viewed by others
Main
In the nervous system, apoptosis has an essential role during development. Accumulating data also suggest that aberrant apoptosis may be involved in some pathological conditions such as stroke and neurodegenerative diseases.1 In neurons, caspases are predominantly activated through the intrinsic pathway, that involves cytochrome c release from mitochondria and formation of the apoptosome, which results in activation of caspase 9 and subsequent activation of downstream caspases.2 The proteins of the Bcl-2 family, that comprises both anti-apoptotic (Bcl-2, Bcl-xL, Mcl-1,…) and pro-apoptotic members (Bax, Bak,…), are key regulators that control the release of cytochrome c and other apoptogenic factors from mitochondria.3 Some signalling pathways regulating the survival/death fate of neurons are now well characterized. For example, the Phosphatidyl Inositol-3-OH Kinase (PI3K)/Akt cascade has an essential role in the survival of many types of neurons,1 notably by inhibiting glycogen synthase kinase 3 (GSK3).4 Little is known, however, regarding the molecular mechanisms that link these signalling pathways to the mitochondrial apoptotic machinery. Increasing data suggest that the ubiquitin–proteasome system (UPS) may constitute one of these links, by regulating the level or the function of key regulatory proteins.5, 6
The UPS is the main non-lysosomal proteolytic system for degrading cellular proteins.7 To be recognized by the proteasome, specific Lys residues in the substrate protein are covalently conjugated to poly-ubiquitin chains in a process that involves the sequential action of E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme) and E3 (ubiquitin protein ligase) enzymes. The E3s confer a high degree of selectivity to this process by recognizing the target proteins.8 Proteasome inhibitors have been shown to prevent apoptosis in several neuronal types.9, 10 This suggests that the ‘road-blockers’ of the apoptotic pathways have to be eliminated by the UPS for the death process to be initiated in neurons.
In a previous study,11 we analyzed the gene expression profile of cerebellar granule neurons (CGN) during early apoptosis. CGN require serum and depolarizing levels of extracellular KCl ([KCl]o=25 mM) for their survival in vitro and die via the mitochondrial apoptotic pathway when deprived of this trophic support.12 CGN apoptosis is dependent on both proteasome activity9 and macromolecular synthesis.12 Among genes highly upregulated in apoptotic versus control CGN, we found Trim17, also known as terf (testis RING finger protein13). Trim17 was first isolated from a rat testis cDNA library13 but it is also expressed in spleen and thymus and to a lesser extent in liver, kidney and brain.14 The protein Trim17 is a member of the TRIM family that is defined by the presence of the tripartite motif (or RBCC), comprising a RING finger domain, one or two B-boxes and an associated coiled-coil domain.15, 16 The tripartite motif can be followed by a variety of C-terminal domains. In Trim17, as in the majority of TRIM proteins,15 it is a PRY-SPRY domain. Recent experimental evidence suggests that the TRIM family represents one of the largest classes of single-protein, RING-containing, E3 ubiquitin ligases.17 Consistently, TRIM17 has been shown to have an E3 activity.14 However, no cellular function has been ascribed to this protein up to now.
In this study, we have examined the role of Trim17 in neuronal apoptosis. We have shown that Trim17 is induced in several models of neuronal apoptosis, both in vitro and in vivo. Moreover, our overexpression and silencing data indicate that Trim17 is necessary and sufficient for inducing the Bax-dependent intrinsic pathway of neuronal apoptosis and that its pro-apoptotic effect depends on its E3 activity. Taken together, our results indicate for the first time that Trim17 is a critical E3 ubiquitin ligase involved in neuronal apoptosis.
Results
Induction of Trim17 mRNA during initiation of neuronal apoptosis
To identify genes whose expression is required for neuronal apoptosis, we used DNA microarrays and we compared gene expression in CGN incubated in serum-free medium containing either 25 mM KCl (K25, survival) or 5 mM KCl (K5, apoptosis) for 4 h.11 Trim17 was one of the most highly upregulated genes after KCl deprivation with a K5/K25 expression ratio of 8.94±1.59 (average±S.D. of three independent experiments). Real-time RT-PCR analysis confirmed this induction with a K5/K25 mRNA ratio of 17.55±4.26. The peak of Trim17 mRNA level, 4 h after KCl deprivation (Figure 1a), precedes the appearance of the first hallmarks of apoptosis.11
Trim17 mRNA is highly increased during transcription-dependent CGN apoptosis. (a) CGN cultures were left untreated (control : time 0) or washed and switched to serum-free medium containing either 25 mM KCl (K25) or 5 mM KCl (K5) for indicated times. Total RNA was extracted and mRNA content for Trim17 was estimated by quantitative RT-PCR. Fold change was calculated by comparison with neurons maintained in initial culture medium (control). Data are means ± S.D. of triplicate determinations and are representative of seven independent experiments. (b) CGN were left untreated or incubated for 4 h in K25 medium in the absence or the presence of 40 μM LY294002 (LY, PI3K inhibitor) or in K5 medium in the absence or the presence of 10 μM AR-A014418 (AR, GSK3 inhibitor). Then mRNA content for Trim17 was estimated under each condition in comparison with control neurons. Data are means ± S.D. from three independent experiments. (c–e) CGN were incubated with different drugs for 24 h and neuronal survival was estimated by an MTT assay. Results are expressed as percentage survival with respect to cultures maintained in K25 medium without drugs. Data are means ± S.D. of triplicate determinations and are representative of three independent experiments. (c) CGN were incubated in K25, K5 or K25 medium supplemented with 40 μM LY294002 (LY), in the absence or presence of 1 μg/ml actinomycin D (ActD, transcription inhibitor). (d) CGN were incubated in K25 or K5 medium in the absence or presence of 10 μM AR-A014418 (AR). (e) CGN were incubated in K25 medium in the absence or presence of 30 μM glutamate (Glu), in the absence or presence of 1 μg/ml actinomycin D
To determine whether the expression of Trim17 was directly related to apoptosis, the Trim17 mRNA levels were assessed in further conditions (Figure 1b). Inhibition of PI3K has been shown to induce transcription-dependent apoptosis in CGN,18 as confirmed in our culture conditions (Figure 1c). Consistently, in CGN incubated with the PI3K inhibitor LY-294002 in K25 medium, Trim17 mRNA was increased to the same extent as in KCl-deprived CGN (Figure 1b). Conversely, inhibition of GSK3 by the very specific inhibitor AR-A01441819 completely prevented CGN death (Figure 1d), confirming previous results with other GSK3 inhibitors.20, 21 In KCl-deprived CGN protected by AR-A014418, induction of Trim17 mRNA was completely abolished (Figure 1b). Collectively, these results demonstrate that Trim17 induction is associated with neuronal apoptosis independently of the [KCl]o, and that expression of Trim17 is controlled by the PI3K/Akt/GSK3 pathway in CGN.
The mRNA level of Trim17 was also significantly increased in two other models of transcription-dependent neuronal apoptosis:22, 23 in rat sympathetic neurons from the superior cervical ganglion (SCG) after NGF withdrawal, and in motoneurons from mouse spinal cord maintained in the absence of neurotrophic factors (Table 1). Although the induction of Trim17 was lower in these models, possibly due to slower apoptotic kinetics, it was reproducible and significant.
To examine whether Trim17 may be part of an adaptive response to stress, we studied its expression during CGN apoptosis induced by low concentrations (30–50 μM) of glutamate. This model of neuronal death is apoptotic (Supplementary Figure S1) but it is transcription-independent,24 as confirmed by our data (Figure 1e). However, the level of Trim17 mRNA did not increase after treatment with glutamate (Table 1). Overall, these observations suggest that Trim17 is specifically expressed in transcription-dependent apoptosis, induced by trophic factor deprivation, but not during transcription-independent apoptosis.
Expression of the Trim17 protein during in vitro and in vivo neuronal apoptosis
To study the expression of Trim17 at the protein level, we generated an anti-peptide polyclonal antibody that we purified by affinity. In western blots, this antibody recognizes overexpressed or recombinant Trim17 very well, but it is not sensitive enough to efficiently detect the endogenous protein. Nevertheless, this antibody generates a weak signal in immunofluorescence that decreases in CGN transduced with a short hairpin RNA that specifically targets Trim17 mRNA (ShRNA Trim17, Figure 8a) and in SCG neurons microinjected with two different small interfering RNAs (siRNAs) against Trim17 (Figure 8d), demonstrating the specificity of immunodetection. We first used this antibody to determine whether Trim17 is also induced at the protein level during neuronal apoptosis. Using confocal microscopy, a faint, but higher Trim17 immunostaining could be detected in KCl-deprived CGN compared to CGN maintained in K25 medium (Figure 2a). A similar increase of the protein level of Trim17 was also detected in SCG neurons after NGF withdrawal (Figure 2b).
Induction of the Trim17 protein during in vitro neuronal apoptosis. (a) Detection of endogenous Trim17 by confocal immunofluorescence using an anti-peptide antibody against Trim17 in primary CGN incubated for 8 h in K25 or K5 medium. The lower pictures show nuclear staining with Hoechst 33258 in the same fields. Note that the most intense Trim17 signals are detected in neurons committed to death but not in neurons already undergoing apoptosis. (b) Detection of endogenous Trim17 by fluorescence microscopy using the same antibody as in panel a in primary SCG neurons incubated for 16 h in the presence or the absence of NGF. The lower pictures show nuclear staining with DAPI in the same fields
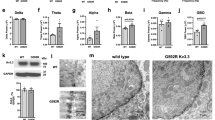
To investigate whether Trim17 is also expressed in vivo, we next performed immunohistochemistry analyses of mouse cerebellum sections. Trim17 immunoreactivity was detected in evenly scattered cells in the external granular layer but more predominantly in the developing internal granular layer, 7 days after birth (P7, Figure 3a, left panel). It is well documented that a peak of naturally occurring neuronal death takes place in these areas at this stage. This death has been demonstrated to be apoptotic25 as confirmed by our immunodetection of active caspase 3 (Figure 3a, right panel). When estimated at different stages of post-natal cerebellar development, Trim17 expression and caspase 3 activation both peaked at P7 (Figure 3b). Trim17 and active caspase 3 could be detected in the same place on adjacent sections (Figure 3c), suggesting that they are present in the same neurons. Taken together, these observations suggest a possible role of Trim17 in developmental neuronal death in vivo.
Expression of the Trim17 protein during developmental neuronal apoptosis. (a) Trim17 and active caspase 3-immunolabelled sagittal cerebellar sections of a post-natal day 7 (P7) mouse. EGL, external granular layer; ML, molecular layer; IGL, internal granular layer. Arrows indicate positive neurons. (b) Sagittal cerebellar sections of mice from different stages of post-natal development were analyzed by immunohistochemistry. The numbers of CGN that were immunopositive for active caspase 3 on one hand, and for Trim17 on the other hand, were estimated and represented per mm2 of cerebellum section using two different scales. (c) Two adjacent sections (4 μm) of cerebellum from a P7 mouse were immunolabelled with antibodies against Trim17 and active caspase 3, respectively. Note the co-localization of the two stainings indicated by an arrow and *(also present in panel a)
E3 ubiquitin-ligase activity of Trim17 and its mutants
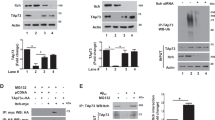
As human TRIM17 has been shown to auto-ubiquitinate in vitro,14 we tested murine Trim17 in similar ubiquitination assays. Immunoblotting shows a single band with increased apparent size after incubation with the E2 enzyme Ube2e1 (Figure 4a, Supplementary Figure S2b), whereas a ladder of higher molecular mass species could be detected with Ube2d2 and with Ube2d3 (Figure 4a and c, Supplementary Figure S2a). These bands likely correspond to mono-ubiquitinated and poly-ubiquitinated forms of Trim17, respectively, as they could be detected by both anti-Trim17 and anti-ubiquitin antibodies. As the reaction mix was free of E3 except Trim17, these results indicate that mouse Trim17 is capable of auto-ubiquitination. In contrast to human TRIM17, which has been shown to preferentially cooperate with Ube2e1 to induce poly-ubiquitination,14 mouse Trim17 also acts efficiently with Ube2d2 and Ube2d3 and generates mostly mono-ubiquitination with Ube2e1 (Figure 4a). These discrepancies may result from differences between human and mouse proteins or between experimental conditions.
E3 ubiquitin-ligase activity of Trim17 and its mutants. (a) Purified recombinant GST-Trim17(WT) was incubated for 2 h in the presence of all in vitro ubiquitination assay components with the exception of ubiquitin and E2 enzyme (first lane), in the presence of ubiquitin and different E2 enzymes (Ube2e1, Ube2d2 and Ube2d3) as indicated. (b) Schematic representation of the WT mouse Trim17 protein and its mutant ΔRING (upper part). Diagram of the RING domain of WT Trim17 and two point mutants of the RING domain: C16A and K2R (lower part). The conserved Cys and His that coordinate two atoms of zinc are represented, as well as the 8 Lys present in the RING. Five of these 8 Lys, that are all gathered within a stretch of 12 residues, are conserved between mouse, rat and human, the three others being replaced by Arg in human. c: In vitro ubiquitination assays were performed using GST-Trim17(WT) and GST-Trim17 mutant proteins (K2R, ΔRING and C16A), in the presence of all of the assay components including Ube2d3 as an E2 enzyme, except in the first lane in which all of the assay components were incubated in the absence of recombinant Trim17 and in the second lane in which GST-Trim17(WT) was incubated in the presence of all assay components but in the absence of ubiquitin. (a and c) The reaction products were analyzed by western blot using a non-purified antibody against full-length Trim17. The membranes were subsequently stripped and reprobed for ubiquitin. The arrows indicate GST-Trim17 and GST-Trim17 (ΔRING). The mono-ubiquitinated form of Trim17 is indicated by **. Higher bands are poly- or multi-ubiquitinated forms of Trim17
To study the cellular function of Trim17, we constructed several RING domain mutants of Trim17 (Figure 4b). The RING is a Cys-rich zinc-binding motif that has been shown to confer E3 ubiquitin ligase activity to many proteins, including most TRIMs.17 Therefore, we first created a deletion (ΔRING) and a point mutation (C16A), in which one of the Cys residues (C16) responsible for coordination of zinc was replaced by Ala (Figure 4b). Consistently, GST-Trim17(ΔRING) and GST-Trim17(C16A) were incapable of auto-ubiquitination in vitro, regardless of the E2 enzyme used (Figure 4c, Supplementary Figure S2). Human TRIM17 has been shown to be degraded by the proteasome when overexpressed in cell lines.14 As mouse Trim17(ΔRING) initially seemed to be more stable than wild-type (WT) Trim17 (data not shown), we hypothesized that Trim17 might be ubiquitinated on its RING domain. In an attempt to create a more stable but still active mutant of Trim17, all the Lys residues present in the RING were replaced by Arg residues (Figure 4b). If Trim17 is actually ubiquitinated on these Lys, this K2R mutant might be less degraded by the proteasome in cells. However, it should maintain the structure of its RING intact. Pulse-chase experiments, however, showed that the half-life of Trim17(K2R) is not significantly different from that of WT Trim17 and of the other Trim17 mutants (Supplementary Figure S3). Nevertheless, in vitro ubiquitination assays showed that Trim17-K2R is capable of auto-ubiquitination (Figure 4c, Supplementary Figure S2a). These data thus indicate that Trim17(WT) and Trim17(K2R) have an E3 ubiquitin-ligase activity, whereas Trim17(ΔRING) and Trim17(C16A) are inactive mutants.
Expression of Trim17 is sufficient to trigger the mitochondrial apoptotic pathway in CGN
To evaluate the effect of Trim17 on neuronal survival/death fate, CGN were transfected with mouse Trim17 fused to green-fluorescent protein (GFP). Expression of Trim17-GFP coincided with chromatin condensation and caspase 3 activation in a large proportion of transfected neurons (Figure 5b). In healthy neurons, the immunostaining pattern for cytochrome c was intense and punctate, indicating a mitochondrial localization, whereas it disappeared in dying neurons expressing Trim17-GFP (Figure 5b). It is likely that this absence of immunoreactivity reflected degradation of cytochrome c following its release from mitochondria, as described elsewhere.26 In contrast, neither cytochrome c release nor activation of caspase 3 was detectable in most CGN transfected with GFP alone or with the inactive mutant Trim17(ΔRING)-GFP (Figure 5a and c). These observations therefore suggest that Trim17 expression can trigger the intrinsic pathway of apoptosis in CGN.
Overexpression of Trim17 induces the intrinsic apoptotic pathway in CGN. CGN were transfected with GFP (a), Trim17-GFP (b) or Trim17(ΔRING)-GFP (c) for 25 h. Then, neurons were fixed and immunostained for cytochrome c or for the active form of caspase 3. Transfected neurons were detected by GFP fluorescence and nuclei were visualized by Hoechst 33258 staining. Thick arrows indicate Trim17-expressing neurons that undergo apoptosis, whereas thin arrows indicate healthy neurons expressing GFP, Trim17-GFP or Trim17(ΔRING)-GFP. In healthy neurons, cytochrome c immunostaining is intense and punctate, indicating a mitochondrial localization. In apoptotic neurons, no staining is visible because cytochrome c is rapidly degraded after release from mitochondria. The shape of GFP-positive neurons was drawn on the cytochrome c picture to facilitate interpretation of the image
To further examine this point, Trim17-GFP was transfected into CGN obtained from Bax-deficient mice and their wild type littermates. Neuronal death has been previously shown to be completely prevented in Bax−/− CGN maintained in low KCl level for several days.27 Similarly, Trim17 did not induce apoptosis in Bax−/− CGN (Figure 6a). Indeed, both WT Trim17–GFP and the active mutant Trim17(K2R)–GFP had a strong pro-apoptotic effect in Bax+/+ and Bax+/− CGN compared to GFP used as a negative control. In contrast, apoptosis was negligible in Bax−/− neurons transfected with the three constructs (Figure 6b). Interestingly, heterozygous Bax+/− CGN died to the same extent as wild-type Bax+/+ CGN upon transfection with Trim17-GFP (Figure 6b), as upon KCl deprivation.27 As Bax is necessary for cytochrome c release, these results demonstrate that Trim17 expression is sufficient to trigger the intrinsic pathway of apoptosis in CGN.
Trim17-induced neuronal apoptosis is Bax-dependent. (a) CGN from WT and Bax-deficient mouse littermates were transfected with Trim17-GFP for 25 h and fixed. Trim17 was detected by GFP fluorescence and nuclei were visualized by Hoechst 33258 staining. Note that the morphology of the nucleus and of the cell body is normal in Bax−/− neurons transfected with Trim17-GFP, whereas the cytoplasm is shrunken and the chromatin condensed or fragmented in a large proportion of Bax+/+ neurons expressing Trim17-GFP (indicated by thick arrows). (b) CGN from Bax−/−, Bax+/− and Bax+/+ mouse littermates were transfected with GFP (control) or with the active forms of Trim17 (WT and K2R) fused to GFP for 25 h. The percentage of apoptosis among transfected neurons was assessed by examining cell morphology and nuclear condensation of GFP-positive neurons. Data are representative of three independent experiments
The E3 ubiquitin-ligase activity of Trim17 is required for neuronal apoptosis
To better assess the role of Trim17 in neuronal apoptosis, the effect of its different mutants was thoroughly quantified in control and apoptotic neurons. First, CGN were transfected with either Trim17-GFP variants or GFP for 16 h, and were maintained in initial culture medium (Ctrl) or switched to apoptotic K5 medium for an additional 8 h period. Then, the percentage of transfected neurons undergoing apoptosis was determined (Figure 7a). Compared to GFP, expression of the active forms of Trim17–GFP (WT and K2R) was sufficient to induce CGN apoptosis in survival conditions, and to aggravate apoptosis after KCl deprivation. In contrast, the inactive mutants Trim17(ΔRING)–GFP and Trim17(C16A)–GFP did not trigger significant apoptosis in control conditions. More importantly, expression of these mutants completely protected CGN from KCl deprivation-induced apoptosis (Figure 7a). This effect was associated with inhibition of cytochrome c release and caspase 3 activation (Figure 7b).
The E3 activity of Trim17 is necessary for neuronal apoptosis. (a) CGN were transfected with GFP (as a negative control) or with different mutants of Trim17 fused to GFP for 16 h and were left untreated (Ctrl) or were incubated in K5 medium for 8 h. Then, CGN were fixed and stained with Hoechst 33258. Among GFP-positive neurons, the percentage of apoptosis was assessed by observing cell morphology and nuclear condensation. Data are the mean ± S.D. of three independent experiments. *P<0.001 significantly different from the corresponding GFP control (ANOVA followed by Bonferroni multiple comparisons test). (b) CGN were transfected with Trim17(ΔRING)-GFP for 16 h and were incubated in K5 medium for 8 h. Then, neurons were fixed and immunostained for cytochrome c or for the active form of caspase 3. CGN-expressing Trim17(ΔRING)-GFP were detected by GFP fluorescence and nuclei were visualized by Hoechst 33258 staining. Arrows indicate neurons expressing Trim17(ΔRING)-GFP. In these neurons, immunostaining for cytochrome c is intense and punctate indicating that it has not been released from mitochondria. In contrast cytochrome c immunostaining is diffuse and fainter in surrounding dying neurons. The shape of GFP-positive neurons was drawn on the cytochrome c picture to facilitate interpretation of the image. (c) SCG neurons were microinjected with the empty vector pCI or plasmids expressing the different forms of Trim17 together with a fluorescent marker. Then, neurons were further cultured in medium containing NGF (+ NGF, only for empty pCI) or lacking NGF (− NGF, for all constructs). Viability of injected SCG neurons was assessed at this time (time 0, 100% survival) and subsequently, every 24 h. Data are the mean ± S.D. of three independent experiments. *P<0.001 significantly different from the value obtained with pCI − NGF at the same time (ANOVA followed by Bonferroni multiple comparisons test)
This strong protection was reproduced in sympathetic SCG neurons deprived of NGF (Figure 7c). In these experiments, expression vectors encoding the different forms of untagged Trim17 were microinjected together with a fluorescent marker. Then, neurons were deprived of NGF and the number of viable injected neurons was determined at this time (0) and subsequently, every 24 h. After NGF withdrawal, SCG neurons injected with the plasmids expressing WT Trim17 and Trim17(K2R) showed a similar time course of neuronal death compared to neurons injected with the empty vector pCI. In contrast, SCG neurons microinjected with expression vectors encoding Trim17(ΔRING) and Trim17(C16A) showed strongly increased survival after NGF deprivation. This protection was close to that observed in neurons cultured in the presence of NGF (Figure 7c) and similar to the effect of Bcl-2 or dominant-negative c-Jun in the same type of microinjection assay.28
Taken together these results show that the pro-apoptotic effect of Trim17 depends on the E3 activity conferred by its RING domain. More importantly, mutations of the RING generally induce a dominant-negative effect.8 Therefore, the protective effect of the ΔRING and C16A mutants strongly suggests that the E3 activity of Trim17 is necessary for neuronal apoptosis both in CGN and SCG neurons.
Knockdown of Trim17 expression in primary neurons prevents apoptosis
To confirm the role of Trim17 in neuronal apoptosis, we knocked down the expression of endogenous Trim17 in CGN by using lentiviruses expressing ShRNAs. One sequence (ShRNA Trim17) that specifically targets the Trim17 mRNA was particularly efficient at decreasing the protein level of Trim17 (Figure 8a). Most importantly, this downregulation was associated with almost complete protection of CGN against KCl deprivation-induced apoptosis. Indeed, nuclear condensation and caspase 3 activation were strongly decreased in CGN transduced with ShRNA Trim17 (Figure 8a–c). In contrast, the control ShRNA, that did not significantly affect the protein level of Trim17 compared to non-transduced neurons (Figure 8a), had no protective effect. It was rather toxic in its own right (Figure 8a, b).
Expression of Trim17 is necessary for neuronal apoptosis. (a) CGN plated on glass coverslips in 24-well plates were not treated (non transduced) or were transduced with lentiviral particles expressing ShRNA sequences (control and Trim17) one day after plating. At DIV 6, neurons were incubated for 8 h in K5 medium. Then, neurons were fixed, immunostained for endogenous Trim17 using the affinity-purified anti-peptide antibody and stained with DAPI to reveal nuclear morphology. The data are representative of two independent experiments. (b) CGN were treated as in panel a and the percentage of apoptotic neurons displaying condensed nuclei was estimated in six random fields per condition (approximately 800 cells counted in total per condition). Data are the mean ±S.D. of four independent experiments. *P<0.01 significantly different from non-transduced neurons in the same condition (ANOVA followed by Bonferroni multiple comparisons test). (c) CGN plated in six-well plates were not treated (non transduced) or were transduced with lentiviral particles expressing ShRNA sequences (control and Trim17) one day after plating. At DIV 6, neurons were incubated for 7 h in K5 medium or maintained in the initial culture medium (ctrl). Then, proteins were extracted and analyzed by western blot using antibodies against caspase 3 and actin. (d) SCG neurons were microinjected with 3 μM siRNA: non-targeting control (NTC), and siRNAs targeting two different sequences in Trim17 mRNA (Trim17 siRNA no. 1 and Trim17 siRNA no. 2) together with guinea pig IgG. After 24 h, neurons were deprived of NGF for 16 h and were fixed. Endogenous Trim17 was detected with the same antibody as in panel a, microinjected neurons were visualized with fluorescent anti-guinea pig secondary antibody and nuclei were stained with DAPI. The data are representative of three independent experiments. The shape of microinjected neurons was drawn on the Trim17 picture to facilitate interpretation of the image. (e) SCG neurons were microinjected with the same siRNAs as in panel d, together with a fluorescent marker. The plasmid expressing Trim17 (pCI-Trim17, 50 ng/μl) was also microinjected as a control to assess the levels of neuronal death in the presence and the absence of NFG. Twenty four hours after microinjection, neurons were further cultured in medium containing NGF (+ NGF, only for pCI-Trim17) or lacking NGF (− NGF, for all other conditions) for 72 h. Viability of injected SCG neurons was then assessed and expressed as percentage of the survival determined at the time of NGF deprivation. Data are the mean ±S.D. of six independent experiments. *P<0.01 significantly different from the value obtained in neurons injected with NTC siRNA (ANOVA followed by Dunnett multiple comparisons test)
Similar results were obtained in SCG neurons microinjected with a non-targeting control siRNA and two siRNAs targeting different specific sequences in Trim17 (siRNA no. 1 and no. 2). Whereas the control siRNA did not affect the level of Trim17 protein significantly, the immunostaining of Trim17 almost completely disappeared in a majority of sympathetic neurons microinjected with siRNA no. 1 and was reduced to a lower, but significant, extent in most neurons injected with siRNA no. 2 (Figure 8d, Table 2). Consistently, survival was significantly increased in SCG neurons microinjected with siRNA no. 1, and slightly improved with siRNA no. 2, compared to neurons injected with the control siRNA, after NGF deprivation (Figure 8e). This suggests that only a profound reduction of the Trim17 protein level can afford protection against death.
To check that the absence of Trim17 is actually responsible for the protection observed after RNA interference, we transfected a form of Trim17 in which the sequence targeted by ShRNA Trim17 has been mutated to be resistant to ShRNA-mediated degradation. Consistently, we could express this form of Trim17 fused to the fluorescent protein mCherry (Trim17-res-mCherry) in CGN previously transduced with ShRNA Trim17 (Figure 9). More importantly, expression of this form of Trim17 restored the ability of neurons to undergo apoptosis after KCl deprivation whereas transfection with mCherry alone did not (Figure 9). Taken together, these silencing data further confirm that Trim17 expression is necessary for the initiation of neuronal apoptosis.
Transfection of a ShRNA-resistant Trim17 construct restores KCl deprivation-induced apoptosis in transduced CGN. One day after plating, CGN were transduced with lentiviral particles expressing the ShRNA sequence directed against Trim17 mRNA that efficiently decreases the level of endogenous Trim17 (ShRNA Trim17). At DIV 5, neurons were transfected with pmCherry-C2 or with pCI-Trim17-res-mCherry (in which the sequence targeted by the ShRNA Trim17 is mutated). At DIV 6, CGN were incubated for 8 h in K5 medium. Then, neurons were fixed, CGN expressing mCherry or Trim17-res-mCherry were detected by mCherry fluorescence and nuclei were visualized by Hoechst 33258 staining. The data are representative of two independent experiments
Discussion
Accumulating evidence indicates that the UPS is involved in the control of apoptosis.5, 6 However, only a few proteins whose degradation by the proteasome is necessary for apoptosis have been identified so far, and the crucial E3 ubiquitin ligases responsible for their recognition remain mostly unknown. In this study, we have identified Trim17 as a critical E3 ubiquitin ligase whose expression is sufficient and necessary for triggering the intrinsic pathway of apoptosis in different neuronal types. Indeed, transfection of CGN with the active forms of Trim17 was sufficient, in itself, to trigger apoptosis in survival conditions. In contrast, two inactive mutants did not induce apoptosis. Moreover, Trim17-induced apoptosis was completely abrogated in Bax−/− CGN that are resistant to KCl deprivation but are still sensitive to glutamate-induced cell death.27 Taken together, these data show that the pro-apoptotic effect of Trim17 is not the toxic consequence of transfection or accumulation of an exogenous protein prone to aggregation, but results from the specific activation of the intrinsic apoptotic pathway. Importantly, the dominant-negative mutants of Trim17 blocked apoptosis both in KCl-deprived CGN and in NGF-deprived SCG sympathetic neurons. Different constructs and different gene transfer methods were used in the two models but gave the same results, underlying the robustness of this observation. This strong protective effect indicates that the E3 activity of Trim17 is necessary for the initiation of neuronal death. This was further confirmed by silencing experiments, using different RNA interference tools, in which the reduction of Trim17 protein afforded a significant protection of both CGN and SCG neurons after trophic factor withdrawal.
By showing that Trim17 has an essential role in the initiation of neuronal apoptosis, our data are the first to assign a cellular function to Trim17. They also establish a novel pathway linking the PI3K/Akt/GSK3 cascade with the mitochondrial apoptotic machinery. In the past years, extensive studies have demonstrated that depolarization-induced calcium entry and activation of neurotrophin receptors can trigger the PI3K/Akt pathway which has a crucial role in neuronal survival,1 notably by inhibiting the pro-apoptotic kinase GSK3β.4 Nevertheless, little is known about the mechanisms that control the apoptotic machinery downstream of Akt or GSK3 in neurons. In CGN, Akt has been shown to phosphorylate the pro-apoptotic BH3-only protein Bad,29 as well as members of the Forkhead family of transcription factors, which suppresses the transactivation of death-promoting genes such as Fas ligand and bim.30, 31 Although the BH3-only protein Bim clearly has an important role,32 analysis of neurons from different KO mice has cast a doubt on the role of Bad and Fas ligand in neuronal apoptosis.33 More recently, phosphorylation of Bax by GSK3β has been shown to promote the translocation of Bax to mitochondria and the induction of apoptosis in CGN.34 By demonstrating that Trim17 has a decisive role in the initiation of neuronal death upstream of Bax and that its expression is controlled by the PI3K/Akt/GSK3 cascade, we have now identified a novel link between these signalling pathways and the mitochondria.
Our immunohistochemistry data indicate that Trim17 protein is expressed in vivo, in the cerebellum, during post-natal development. Its peak of expression coincides with the peak of naturally occurring neuronal apoptosis that takes place at P7. It is thus plausible that the role of Trim17, that we have identified in vitro, is also of crucial importance for apoptosis in vivo. Moreover, the fact that Trim17 is highly expressed in organs with a high rate of constitutive apoptosis, such as testis, spleen and thymus,14 is consistent with a possible role of Trim17 in the regulation of apoptosis in other cell types.
In this study, we present evidence that Trim17 has an essential role in the triggering of neuronal apoptosis via its E3 activity. Our work is therefore a new contribution to the picture of apoptosis regulation by the UPS and provides a new starting point to elucidate these mechanisms. A simple hypothesis would be that, following an apoptotic stimulus, de novo expressed Trim17 ubiquitinates and targets key anti-apoptotic proteins to the proteasome. Identification of ubiquitination substrates of Trim17 is thus under progress. However, ubiquitination mediated by Trim17 may have other consequences than proteasomal degradation: change of localization, activation or neutralization, change in expression. Moreover, TRIM proteins often associate in homo/hetero-oligomers through their coiled-coil domains to form large protein complexes.15 For example, TRIM17 has been shown to interact with, and to be stabilized by TRIM44.14 It is thus plausible that the activity of Trim17 may be influenced or may influence other members of the TRIM family. This will have to be taken into consideration for the identification and validation of Trim17 substrates.
Materials and Methods
Primary cultures of neurons
Bax+/− mice were generated by the laboratory of Dr. S Korsmeyer35 and were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) (strain B6.129X1-Baxtm1Sjk/J). Bax-deficient mice (Bax−/−) were obtained by mating of heterozygotes. The offspring were genotyped by PCR according to the protocol provided by The Jackson Laboratory, with genomic DNA prepared from tails using the REDExtract-N-Amp Tissue PCR Kit (Sigma-Aldrich, Saint Quentin Fallavier, France). Common C57Bl/6 mice (from Charles River Laboratories, Saint Germain sur l'Arbresle, France) were bred in house.
CGN cultures were prepared from 7-day-old murine pups as described previously11 with slight modifications. In brief, freshly dissected cerebella were incubated for 10 min at 37°C with 0.25 mg/ml trypsin and cells were dissociated in HBSS −Ca2+ −Mg2+ in the presence of 0.5 mg/ml trypsin inhibitor and 0.1 mg/ml DNaseI by several steps of mechanical disruption with a flame-polished Pasteur pipette. The resulting cell suspension was filtered through a 40-μm cell strainer (BD Falcon, Le Pont de Claix, France) and centrifuged at 200 × g for 10 min. Cells were gently resuspended in K25+S medium (Basal Medium Eagle (BME, GIBCO Invitrogen, Cergy Pontoise, France) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 10 mM HEPES, penicillin–streptomycin 100 IU/ml–100 μg/ml and 20 mM KCl to achieve a final concentration of 25 mM), counted and seeded at a density of 2 50 000 cells/cm2 in culture dishes previously coated with 20 μg/ml poly-D-lysine (Becton Dickinson Biosciences, Le Pont de Claix, France). For immunofluorescence and transfection experiments, CGN were cultured on glass coverslips coated with 16.67 μg/ml laminin (Sigma-Aldrich) and 33.3 μg/ml poly-D-lysine. CGN were cultured at 37°C in a humidified incubator with 5% CO2/95% air for 6 days. To prevent proliferation of remaining non-neuronal cells, the anti-mitotic cytosine β-D-arabinofuranoside (10 μM Ara-C) was added to the culture medium 24 h after plating. At 6 days in vitro (DIV) granule neurons represented more than 98% of cultured cells (data not shown).
Sympathetic neurons were isolated from the superior cervical ganglia (SCG) of one-day-old Sprague–Dawley rats and cultured as previously described.36 The ganglia were dissociated by incubation for 30 min at 37°C in 0.025% trypsin and then for 30 min at 37°C in 0.1% collagenase, followed by trituration through the tip of a Gilson P1000. After centrifugation (at 200 × g for 10 min), neurons were gently resuspended in SCG medium (DMEM containing 4.5 g/l glucose and 110 mg/l pyruvate (Sigma-Aldrich) supplemented with 10% fetal calf serum, penicillin/streptomycin and 2 mM L-glutamine) containing 50 ng/ml 2.5S NGF (Cedarlane, Burlington, ON, Canada) to sustain survival, and fluorodeoxyuridine (20 μM) and uridine (20 μM) to prevent the proliferation of non-neuronal cells. Typically 6400–8000 neurons were plated on 13 mm diameter glass coverslips coated with poly-L-lysine and laminin. Fresh medium was added after 1, 2, 4 and 6 days in culture. Cells were used for experiments at DIV 5–7. In NGF-withdrawal experiments, neurons were rinsed twice with medium (without NGF) and were re-fed with a medium containing anti-NGF antibody (100 ng/ml).
Motoneuron cultures were prepared from E12.5 ventral mouse spinal cords as described previously37 with slight modifications. In brief, cells were dissociated after trypsin treatment. The largest cells were isolated by centrifugation on a 5.2% Optiprep density gradient and were subsequently centrifuged through a bovine serum albumin cushion. Motoneurons were resuspended in neurobasal medium supplemented with 2% horse serum, 25 μM L-glutamate, 25 μM β-mercaptoethanol, 0.5 mM L-glutamine and 2% B-27 supplement (GIBCO Invitrogen). Motoneurons were plated on plates previously coated with polyornithine/laminin, in the presence or not of a cocktail of neurotrophic factors consisting of 1 ng/ml human brain-derived neurotrophic factor (BDNF), 100 pg/ml rat glial cell line-derived neurotrophic factor and 10 ng/ml ciliary neurotrophic factor, added at the time of seeding.
RNA preparation and real time quantitative RT-PCR
Total RNA was extracted using the RNAqueous kit (Ambion, Applied Biosystems, Les Ulisses, France) and treated with DNase I from the DNA-free kit (Ambion) according to the manufacturer's instructions. RNAs were used to perform a two-step reverse-transcription polymerase chain reaction (RT-PCR). In brief, 1 μg of total RNA was reverse transcribed using 200 U M-MLV reverse transcriptase (Invitrogen) in the presence of 2.5 μM N6 random primers and 0.5 mM dNTP. The equivalent of 6 ng of resulting cDNA was used as a template for real time PCR using a M × 3000P thermocycler (Agilent Technologies, Massy, France) with a home-made SYBR Green QPCR master mix.38 The sequences of the primers used were the following: for mouse Trim17: 5′ AGGGAGTATAAGCTCAAGTTGGA 3′ and 5′ CCTGCCACTCAGTTAAGGTCT 3′; for mouse cJun: 5′ GCCCTCAACGCCTCGTT 3′ and 5′ GCCAGGTTCAAGGTCATGCT 3′; for rat Trim17: 5′ CTACCCAATTCCATTCCAGTCACAC 3′ and 5′ CAAGCAGAAGAACGGCAGCAG 3′; for rat cJun: 5′ GTGAAGTGACCGACTGTTCTATGAC 3′ and 5′ AATCTTAGGGTTACTGTAGCCGTAGG 3′. PCRs were performed in 10 μl in the presence of 150–300 nM primers. Thermal cycling parameters were 10 min at 95°C, followed by 40 cycles of 95°C for 30 s, 64°C for 30 s and 72°C for 30 s. Data were analyzed and relative amounts of specifically amplified cDNA were calculated with MxPro software (Agilent). The mouse and the rat β-2 Microglobulin amplicons were used as references (mouse primers: 5′ TATGCTATCCAGAAAACCCCTCAA 3′ and 5′ GTATGTTCGGCTTCCCATTCTC 3′; rat primers: 5′ TCTTTCTGGTGCTTGTCTCTCTGG 3′ and 5′ CATCGGTCTCGGTGGGTGTG 3′).
KCl deprivation of CGN cultures and survival assay
At DIV 6, CGN cultured in 24 well plates were washed once and then incubated for indicated times in serum-free BME supplemented with L-Gln, HEPES, antibiotics and 1 μM of the NMDA antagonist (+)-MK-801, and containing either 25 mM KCl (K25 medium) or 5 mM KCl (K5 medium) to respectively sustain survival or induce apoptosis. When indicated, different drugs were added to K25 and K5 media. LY-294002 and AR-A014418 (GSK3 inhibitor VIII) were from Calbiochem (Merck chemicals, Nottingham, UK) and other chemicals were from Sigma-Aldrich. In the case of glutamate treatment, (+)-MK-801 was omitted. Then, neuronal survival was assessed in triplicate by the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazoliumbromide) assay. Briefly, culture medium was replaced by K25 medium containing 0.5 mg/ml MTT and neurons were returned to the incubator for 2 h. Medium was removed and blue formazan produced by living cells was dissolved in DMSO. The optical density of blue formazan was estimated at 570 nm.
cDNA cloning of Trim17 and generation of specific antibodies
Single-stranded random-primed cDNA prepared from mouse apoptotic CGN was used as a template for PCR amplification of Trim17 cDNA using the following primers 5′ GGAATTCTGAGCCATGGATGCGGTGGAGCTTG 3′ and 5′ GCTCTAGACTATCCCTTCACCCACATGGTC 3′. The PCR product was cloned between the EcoRI and XbaI sites of pCI (Promega, Charbonnières, France) to create pCI-Trim17. To produce recombinant Trim17 in Escherichia coli, an 8-histidine tag was first added to the N-terminus of Trim17 by annealing oligonucleotides 5′ CTAGCCATGGCGCATCATCACCATCATCACCATCACGGCGCGCCG 3′ and 5′ AATTCGGCGCGCCGTGATGGTGATGATGGTGATGATGCGCCATGG 3′ and cloning them between the NheI and EcoR1 sites of pCI-Trim17 to create pCI-His(8)-Trim17. An EcoR1-SalI fragment was sub-cloned from this plasmid into pET-23d (+) plasmid (Novagen, Merck chemicals) to create pET-23d-His(8)-Trim17.
To raise a polyclonal antibody against recombinant full-length Trim17, pET-23d-His(8)-Trim17 was transformed in Rosetta (DE3) Escherichia coli. Protein expression was induced by 400 μM of isopropyl β-D-thiogalactopyranoside (IPTG) and was carried out at 25°C for 4 h. Then, bacteria were lysed in Bugbuster HT (Novagen). Inclusion bodies containing Trim17 were dissolved in Laemmli buffer and Trim17 was separated by preparative SDS PAGE. The band containing Trim17 was excised and electro-eluted in 500 μl buffer (Tris 25 mM, Gly 192 mM, SDS 0.025%). About 10 μg of protein was injected to immunize rabbits. Another rabbit polyclonal antibody was generated by Eurogentec (Seraing, Belgium) against two peptides from Trim17: H2N-RQRRYLSPPPEGSAPY-CONH2 (peptide 339) and H2N-KSFQEDVMPDPSSAC-CONH2 (peptide 340) following the double X program (immunization with both peptides at the same time). Sera were affinity-purified using peptide 340 immobilized on HiTrap NHS-activated columns (Amersham, GE Healthcare Europe, Orsay, France) according to the manufacturer's instructions.
Immunofluorescence
After apoptosis induction, or 1 day after transfection, CGN were fixed with 4% paraformaldehyde for 20 min at room temperature. They were then washed with PBS and permeabilized with 0.5% Triton X-100 in PBS at room temperature for 5 min. After blocking with PBS + 5% normal goat serum for 30 min at room temperature, coverslips were incubated overnight with primary antibodies diluted in PBS + 5% normal goat serum at 4°C: mouse monoclonal antibody against the native form of cytochrome c (1 : 20; BD Pharmingen no. 556432, Le Pont de Claix, France), rabbit polyclonal antibody against the cleaved form (Asp175) of caspase 3 (1 : 500; Cell Signaling Technology no. 9661, Beverly, MA, USA) or purified anti-peptide antibody against Trim17 (1 : 100). Neurons were then washed twice in PBS and developed with Alexa Fluor 488 and Alexa Fluor 594-labeled goat anti-mouse and goat anti-rabbit antibodies (Molecular Probes, Invitrogen, dilution 1 : 1000 in PBS + 5% normal goat serum). In the last wash, 1 μg/ml of Hoechst 33258 (Sigma) was added to the cells for 10 min at room temperature. Coverslips were set in Mowiol (polyvinyl alcohol 4–88, Fluka) on glass slides and analyzed for fluorescence. SCG neurons were treated as CGN, except that blocking was performed using 50% normal goat serum and 1% bovine serum albumin (BSA) in PBS and the anti-Trim17 antibody (diluted in 10% normal goat serum and 1% BSA in PBS) was incubated for 90 min at room temperature. Two secondary antibodies (Jackson Immunoresearch Inc., Suffolk, UK) were used: a fluorescein-conjugated goat anti-rabbit antibody (1 : 500) to detect Trim17 and a rhodamine-conjugated donkey anti-guinea pig antibody (1 : 100) to detect microinjected neurons. The nuclei of SCG neurons were stained with DAPI dye in Antifade (Dako) and the neurons were mounted on glass slides.
Immunohistochemistry
Mouse brains from different stages of post-natal development were isolated and fixed in 4% paraformaldehyde in PBS overnight at 4°C, dehydrated and embedded in paraffin. Sagittal sections (4-μm thick) were cut, mounted on slides, incubated in citrate buffer for 30 min at 99°C for antigen retrieval, incubated in 1% H2O2 for 10 min at room temperature to inhibit endogenous peroxidases, blocked with 20% normal horse serum in TBS for 30 min and with avidin and biotin for 15 min (Blocking kit, Vector laboratories, Burlingame, CA, USA), with three washes in TBS plus 0.1% Tween 20 (TBST) after each step. Then, slides were incubated for 2 h at room temperature in a 1 : 50 dilution of purified anti-peptide Trim17 antibody or 1 : 100 cleaved caspase 3 antibody in TBS plus 5% horse serum. After TBST washes, the slides were incubated with biotinylated goat anti-rabbit secondary antibody (Vector laboratories) in TBS plus 5% horse serum, for 45 min at room temperature. After TBST washes, the immunolabeling was revealed using the Vectastain ABC kit and DAB (Vector laboratories) according to the manufacturer's instructions. Following dehydration, slides were mounted in Vectamount medium (Vector laboratories) and analyzed by conventional microscopy.
Site-directed mutagenesis of Trim17
The pCI-Trim17(C16A) mutant was obtained by site-directed mutagenesis of pCI-Trim17, using the QuikChange II XL kit (Agilent Technologies), with the primers 5′ CAAGAGGAGGCCACAGCTTCCATCTGCCTTGACTAC 3′ and 5′ GTAGTCAAGGCAGATGGAAGCTGTGGCCTCCTCTTG 3′. pCI-Trim17(ΔRING) was obtained by removing an EcoR1-StuI fragment from pCI-Trim17 and replacing it with an EcoR1-StuI fragment (lacking the RING domain) cloned by PCR with primers 5′ TTAGAATTCTCCCCTCAGAGGAACCTGAGGC 3′ and 5′ CAGGGAGGCCTTGCTGTCCTG 3′, using pCI-Trim17 as template. The pCI-Trim17(K2R) mutant, in which the eight lysine residues of the RING domain were replaced by arginines, was obtained by recombinant PCR. Two overlapping fragments were amplified using pCI-Trim17 as template, the first with primers 5′ CGAGAATTCTGAGCCATGGATGC 3′ and 5′ CCTCTGCCTCCTCCTCCCCCTTCGAACCCTCCCCCTCTCCCAGCTCATCTGGATGC 3′, the second with primers 5′ AGCTGGGAGAGGGGGAGGGTTCGAAGGGGGAGGAGGAGGCAGAGGGCTCCTTCCCCTGCCC 3′ and 5′ CAGGGAGGCCTTGCTGTCCTG 3′. The two amplicons were purified on a 1% agarose gel. Ten nanograms of each was mixed and used as template for a third PCR amplification using primers 5′ CGAGAATTCTGAGCCATGGATGC 3′ and 5′ CAGGGAGGCCTTGCTGTCCTG 3′. This created a mutated EcoR1-StuI fragment that was used to replace the WT EcoR1-StuI fragment in pCI-Trim17 to give pCI-Trim17(K2R). GFP was fused to the C terminus of Trim17 in pCI by recombinant PCR. A Trim17 fragment with an EGFP overhang, and an EGFP fragment with a Trim17 overhang were amplified by PCR using pCI-Trim17 and pEGFP (Clontech, Saint Germain en Laye, France) as templates and primer pairs 5′ CGAGAATTCTGAGCCATGGATGC 3′/5′ GCCCTTGCTCACGGATCCTCCCTTCACCCACATGGTCACTG 3′ and 5′ CAGTGACCATGTGGGTGAAGGGAGGATCCGTGAGCAAGGGC 3′/5′ AAATCTAGACTATTACTTGTACAGCTCGTCCATG 3′ respectively. The two amplicons were purified on a 1% agarose gel. Ten ng of each was mixed and used as template for a third PCR amplification using primers 5′ CGAGAATTCTGAGCCATGGATGC 3′ and 5′ AAATCTAGACTATTACTTGTACAGCTCGTCCATG 3′. The resulting EcoR1-XbaI fragment was cloned into pCI, to create pCI-Trim17-GFP. pCI-Trim17(C16A)-GFP, pCI-Trim17(ΔRING)-GFP and pCI-Trim17(K2R)-GFP were created by substituting the EcoR1-StuI fragment of pCI-Trim17-GFP with those of the respective pCI-Trim17 mutants. The ShRNA-resistant Trim17 mutant was obtained by recombinant PCR. Two overlapping fragments were amplified using pCI-Trim17 as template, the first with primers 5′ CGAGAATTCTGAGCCATGGATGC 3′ and 5′ CAATGTACTGTTTGGAAGTAGGGACAGCTGTTTTTTCC 3′, the second with primers 5′ CTACTTCCAAACAGTACATTGGTCACTCTCACAGAGCC 3′ and 5′ GACTCTAGACTATCCCTTCACCCACATGG3′. The two amplicons were purified on a 1% agarose gel. Ten ng of each were mixed and used as template for a third PCR amplification using primers 5′ CGAGAATTCTGAGCCATGGATGC 3′ and 5′ GACTCTAGACTATCCCTTCACCCACATGG 3′. The resulting amplicon, in which the ShRNA target site was mutated from 5′ ctgttacccaattccactcta 3′–5′ ctacttccaaacagtacattg 3′ (same amino acids encoded by different codons), was cloned between the EcoR1 and XbaI sites of pCI, to create pCI-Trim17-res. pCI-Trim17-res-mCherry was created by recombinant PCR. Two overlapping fragments were amplified, the first using pCI-Trim17-res as template, with primers 5′ CGAGAATTCTGAGCCATGGATGC 3′ and 5′ CTCCTCGCCCTTGCTCACTCCCTTCACCCACATGG 3′. The second PCR used pmCherry-C2 (Clontech) as template and primers 5′ CCATGTGGGTGAAGGGAGTGAGCAAGGGCGAGGAG 3′ and 5′ ATATCTAGATTACTTGTACAGCTCGTCCATG 3′. The two amplicons were purified on a 1% agarose gel. Ten ng of each were mixed and used as template for a third PCR amplification using primers 5′ CGAGAATTCTGAGCCATGGATGC 3′ and 5′ ATATCTAGATTACTTGTACAGCTCGTCCATG 3′. The resulting amplicon was cloned between the EcoR1 and XbaI sites of pCI, to create pCI-Trim17-res-mCherry. The sequences of all the constructs of Trim17 were confirmed by sequencing.
Production of GST-Trim17 recombinant proteins and in vitro ubiquitination assay
To produce recombinant GST-Trim17 fusion proteins in Escherichia coli, an N-terminal GST-tagged Trim17 was produced by PCR amplification of the Trim17 coding region using pCI-Trim17 or the different pCI-Trim17 mutants as templates and primers 5′ CGAGAATTCGATGCGGTGGAGC 3′ and 5′ CGACTCGAGCTATCCCTTCACC 3′. The PCR products were cloned between the EcoRI and XhoI sites of the pGEX4T1 vector (Amersham, GE Healthcare). The resulting pGEX-Trim17 and pGEX-Trim17 mutants were transformed in BL21 codon plus (DE3) RP Escherichia coli. Protein expression was induced by the addition of 200 μM IPTG and was carried out at 20°C for 4 h in the presence of 100 μM ZnCl2 and 200 μM MgSO4. Bacteria were harvested by centrifugation and resuspended in TNGN buffer (50 mM Tris-HCl pH =7.5, 100 mM NaCl, 10% glycerol, 1% NP-40, 100 μM ZnCl2, 200 μM MgSO4) supplemented with fresh DTT 1 mM and protease inhibitor cocktail (complete EDTA-free, Roche, Mannheim, Germany). Bacterial suspensions were incubated for 30 min at 4°C with 1 mg/ml lysozyme (Fluka) and lysed by sonication. The soluble protein fraction was recovered by centrifugation. GST fusion proteins were isolated by binding to glutathione magnetic beads (MagneGST Glutathione Particles, Promega) pre-equilibrated with PBS. The soluble protein fraction was allowed to bind to the beads for 30 min at room temperature on a rotating wheel before washing the beads three times with 0.65 M NaCl and three times with PBS. In vitro ubiquitination reactions were carried out by adding 50 ng of human recombinant His-tagged ubiquitin-activating enzyme E1 (from BostonBiochem no. E-304, Cambridge, MA, USA), 500 ng human recombinant His-tagged ubiquitin-conjugating enzyme (E2) Ube2e1 (from Biomol International, Enzo Life Sciences AG, Lausen, Switzerland) or purified Ube2d2 or Ube2d3, and 10 μg of ubiquitin (Sigma) to ≈1 μg of recombinant GST-Trim17 still bound to the beads in 20 μl of ubiquitination assay buffer (50 mM Tris-HCl pH =7.5, 50 mM NaCl, 4 mM ATP, 4 mM MgCl2, 2 mM DTT, 10 mM phosphocreatine, 0.5 U creatine kinase, 20 μM ZnCl2. When indicated, the E2 was replaced by 1 μl of wheat germ extract (Promega). Reactions were incubated at 37°C for 2 h under gentle shaking, and then stopped by adding 10 μl of 2 × SDS-PAGE loading buffer and heating at 95°C for 5 min. The samples were immediately resolved by SDS-PAGE and blotted for ubiquitinated protein products using the non purified antibody against full-length Trim17. Membranes were subsequently stripped and reprobed with a rabbit polyclonal anti-ubiquitin antibody (Dako no. Z0458).
Transfection of CGN
CGN grown on glass coverslips in 24-well plates were transfected at DIV 6 with pCI expression vectors encoding GFP (as a control) or different forms of Trim17 fused to GFP. For this purpose, we used a calcium phosphate protocol that has been optimized for neuronal cultures by the group of Dr. M E Greenberg,39 with slight modifications. Briefly, the conditioned culture medium was removed, filtered and saved. The neurons were washed twice and incubated for 1 h with DMEM (Dulbecco's modified Eagle's medium) supplemented with 25 mM KCl, 1 μM (+)-MK-801, 10 mM MgCl2 and 5 mM HEPES, pH =7.5. During this time, the DNA/calcium phosphate precipitate was prepared by mixing one volume of DNA in 250 mM CaCl2 with an equal volume of 2 × HBS (274 mM NaCl, 10 mM KCl, 1.4 mM Na2HPO4, 15 mM D-glucose, 42 mM HEPES, pH 7.0). Two μg of plasmid DNA in 30 μl of CaCl2, mixed with 30 μl 2 × HBS, was used per well. The precipitate was allowed to form for 30 min at room temperature. Then, 60 μl of the DNA/calcium phosphate precipitate were added dropwise to each well and cultures were returned to the CO2 incubator for 50–60 min. The incubation was stopped by ‘shocking’ neurons for 1 min with 1 × HBS supplemented with 10 mM MgCl2, 2% DMSO and 5 mM HEPES, pH =7.5. CGN were then washed twice with DMEM supplemented with 25 mM KCl and 1 μM (+)-MK-801. The saved conditioned medium was added back to each well and the cultures were returned to the 5% CO2 incubator at 37°C. This protocol allows a transfection efficiency of up to 15% with minimal toxicity. Sixteen hours after transfection, CGN were incubated in K5 medium for 8 h or left untreated. CGN were fixed, stained with Hoechst 33258 and analyzed by immunofluorescence in some experiments. Coverslips were mounted on glass slides in Mowiol and transfected neurons were detected by GFP fluorescence. Among GFP-positive neurons, apoptosis was assessed by observing the morphology of the cell and detection of nuclear condensation or fragmentation by Hoechst staining. For each experiment and each condition, at least 200 GFP-positive neurons were scored in a blinded manner. Statistical analyses were performed using GraphPad InStat version 3.0 for Mac (GraphPad Software, San Diego California USA, www.graphpad.com).
Microinjection of SCG sympathetic neurons
SCG sympathetic neurons were microinjected with expression vectors (50 ng/μl) together with 5 μg/μl Texas Red dextran Mr=70 000 (Molecular Probes, Invitrogen) as a marker in 0.5 × PBS (−Ca2+; −Mg2+) as described previously.31 A few hours after injection, the neurons were rinsed twice with fresh SCG medium lacking NGF and further cultured with fresh SCG medium containing NGF (+ NGF) or anti-NGF antibody (− NGF). The injected cells were then counted using an inverted fluorescence microscope and the number of viable, morphologically normal injected (Texas Red positive) neurons at this time (0 h) was set to 100% in each case. Viable injected SCG neurons were recounted 24, 48, 72 and 96 h after NGF withdrawal in a blinded manner. Three independent experiments were performed with 120−140 neurons injected per injection mix.
Trim17 siRNAs, or non-targeting control siRNA, were microinjected as described previously.40 3 μM of the individual siRNA were microinjected into the nucleus together with 2.5 μg/μl of purified guinea-pig IgG (Jackson Immunoresearch Inc.) as a marker. Microinjected cells were returned to culture for 24 h to allow the pre-existing Trim17 protein to degrade. Then, neurons were deprived of NGF for 16 h, fixed and stained for immunofluorescence analysis. Alternatively, neurons were microinjected with siRNAs together with a fluorescent marker, cultured for 24 h and deprived of NGF for increasing times. Viability was assessed as described above. Non-targeting control siRNA oligonucleotides (Dharmacon accession number D-001810–03–05) were purchased from Dharmacon (Lafayette, CO, USA). The sequences targeted by the Trim17 siRNAs used were the following: Trim17 no. 1 5′ UGUUUCUGGUGGAAGAAGA 3′; Trim17 no. 2 5′ ACUGGGAAGUGGGCAUGAA 3′.
Lentivirus preparation and lentiviral transduction of CGN
The HIV-derived lentiviral vectors pLKO.1 containing the ShRNAs TRCN0000037335 (ShRNA control) and TRCN0000037337 (ShRNA Trim17) were obtained from Open Biosystems. They both target specific sequences in Trim17 mRNA but TRCN0000037335 appeared to be inefficient in reducing the level of endogenous Trim17 protein and was thus used as a negative control. Lentiviruses were produced as described.41 Briefly, Lenti-X 293T cells (Clontech) were transfected with pLKO. 1-derived constructs together with the pCMVΔR8.91 and phCMV-G (expressing the vesicular stomatitis G envelope) packaging plasmids at a 3 : 3 : 1 ratio using calcium phosphate precipitation. One day after transfection, the initial culture medium was replaced by a small volume of serum-free medium for 24–30 h. The conditioned medium was harvested, passed through a 0.45 μm pore filter and concentrated by ultracentrifugation using an SW28 rotor (Beckman) at 80 000 × g for 2 h at 4°C. The lentiviruses were resuspended in serum-free medium and titrated using a p24 ELISA kit (Innotest from Innogenetics, Ghent, Belgium) according to the instructions of the manufacturer. Lentiviral preparations were kept frozen in small aliquots at −80°C until use. CGN were transduced 1 day after plating, in the absence of Ara-C. Approximately 500 ng p24 of each lentiviral preparation were added, per million neurons, directly to the culture medium (in a reduced volume) for 8 h. Neurons were then replaced in fresh medium and culture was continued until 6 DIV.
Western blot analysis
Following transduction with lentiviruses expressing different ShRNAs for 5 days and KCl deprivation for 7 h, CGN were harvested in lysis buffer (50 mM Tris-HCl (pH 7.5), 250 mM NaCl, 0.1% Triton X-100, 1 mM DTT, 5 mM iodoacetamide, 20 μM MG-132 and protease inhibitor cocktail (Sigma)) and homogenized by thorough vortexing. Cell debris were removed by centrifugation at 500 × g for 5 min at 4°C, and the protein concentration of the resulting supernatant was estimated using the Bradford reagent (Sigma) with BSA as the standard. Aliquots (15 μg) of supernatant protein were diluted in sample buffer, boiled for 5 min, separated by 15% SDS-PAGE and finally transferred to Hybond-P (Amersham, GE Healthcare) PVDF membrane. After blocking non-specific sites for 1 h at room temperature with 5% non-fat milk in Tris-buffered saline (TBS) supplemented with 0.1% Tween-20, the membrane was incubated overnight at 4°C with primary antibodies diluted in TBS-Tween supplemented with 2.5% milk: anti-cleaved caspase 3 (1 : 1000) and polyclonal anti-caspase 3 (Stressgen no. AAP-103; 1 : 500, Assay Designs, Ann Arbor, MI, USA). After washing the membranes three times in TBS-Tween, the blots were incubated with the anti-rabbit horse radish peroxidase-linked goat secondary antibody (Jackson ImmunoResearch Laboratories Inc.) diluted in TBS-Tween supplemented with 2.5% milk for 1 h at room temperature and subsequently washed three times in TBS-Tween. The immunoreactive proteins were visualized using enhanced chemiluminescent substrate (Pierce, Thermo Fisher Scientific, Rockford, IL, USA). To confirm equal loading and transfer, the membrane was subsequently stripped and reprobed for actin (Chemicon International, Millipore, Billerica, MA, USA, mouse monoclonal antibody no. MAB1501R; 1 : 2000).
Accession codes
Abbreviations
- UPS:
-
ubiquitin–proteasome system
- CGN:
-
cerebellar granule neurons
- [KCl]o:
-
extracellular concentration of KCl
- K25:
-
serum-free medium containing 25 mM KCl
- K5:
-
serum-free medium containing 5 mM KCl
- PI3K:
-
Phosphatidyl Inositol-3-OH Kinase
- GSK3:
-
Glycogen Synthase Kinase 3
- SCG:
-
superior cervical ganglion
- NGF:
-
nerve-growth factor
- ML:
-
molecular layer
- P:
-
days after birth (post-natal)
- E:
-
days of embryonic development
- DIV:
-
days in vitro
- WT:
-
wild type
- GFP:
-
green-fluorescent protein
- ShRNA:
-
short hairpin RNA
- siRNA:
-
small interfering RNA
- BSA:
-
bovine serum albumin
References
Yuan J, Yankner BA . Apoptosis in the nervous system. Nature 2000; 407: 802–809.
Riedl SJ, Salvesen GS . The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol 2007; 8: 405–413.
Youle RJ, Strasser A . The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008; 9: 47–59.
Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA . Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 1995; 378: 785–789.
Broemer M, Meier P . Ubiquitin-mediated regulation of apoptosis. Trends Cell Biol 2009; 19: 130–140.
Thompson SJ, Loftus LT, Ashley MD, Meller R . Ubiquitin-proteasome system as a modulator of cell fate. Curr Opin Pharmacol 2008; 8: 90–95.
Glickman MH, Ciechanover A . The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002; 82: 373–428.
Pickart CM . Mechanisms underlying ubiquitination. Annu Rev Biochem 2001; 70: 503–533.
Canu N, Barbato C, Ciotti MT, Serafino A, Dus L, Calissano P . Proteasome involvement and accumulation of ubiquitinated proteins in cerebellar granule neurons undergoing apoptosis. J Neurosci 2000; 20: 589–599.
Sadoul R, Fernandez P-A, Quiquerez A-L, Martinou I, Maki M, Schröter M et al. Involvement of the proteasome in the programmed cell death of NGF-deprived sympathetic neurons. EMBO J 1996; 15: 3845–3852.
Desagher S, Severac D, Lipkin A, Bernis C, Ritchie W, Le Digarcher A et al. Genes regulated in neurons undergoing transcription-dependent apoptosis belong to signaling pathways rather than the apoptotic machinery. J Biol Chem 2005; 280: 5693–5702.
D'Mello SR, Galli C, Ciotti T, Calissano P . Induction of apoptosis in cerebellar granule neurons by low potassium: inhibition of death by insulin-like growth factor I and cAMP. Proc Natl Acad Sci USA 1993; 90: 10989–10993.
Ogawa S, Goto W, Orimo A, Hosoi T, Ouchi Y, Muramatsu M et al. Molecular cloning of a novel RING finger-B box-coiled coil (RBCC) protein, terf, expressed in the testis. Biochem Biophys Res Commun 1998; 251: 515–519.
Urano T, Usui T, Takeda S, Ikeda K, Okada A, Ishida Y et al. TRIM44 interacts with and stabilizes terf, a TRIM ubiquitin E3 ligase. Biochem Biophys Res Commun 2009; 383: 263–268.
Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L et al. The tripartite motif family identifies cell compartments. EMBO J 2001; 20: 2140–2151.
Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G . Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol 2008; 8: 225.
Meroni G, Diez-Roux G . TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 2005; 27: 1147–1157.
Miller TM, Tansey MG, Johnson EMJ, Creedon DJ . Inhibition of phosphatidylinositol 3-kinase activity blocks depolarization- and insulin-like growth factor 1-mediated survival of cerebellar granule cells. J Biol Chem 1997; 272: 9847–9853.
Bhat R, Xue Y, Berg S, Hellberg S, Ormo M, Nilsson Y et al. Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418. J Biol Chem 2003; 278: 45937–45945.
Cross DA, Culbert AA, Chalmers KA, Facci L, Skaper SD, Reith AD . Selective small-molecule inhibitors of glycogen synthase kinase-3 activity protect primary neurones from death. J Neurochem 2001; 77: 94–102.
Hongisto V, Smeds N, Brecht S, Herdegen T, Courtney MJ, Coffey ET . Lithium blocks the c-Jun stress response and protects neurons via its action on glycogen synthase kinase 3. Mol Cell Biol 2003; 23: 6027–6036.
Comella JX, Sanz-Rodriguez C, Aldea M, Esquerda JE . Skeletal muscle-derived trophic factors prevent motoneurons from entering an active cell death program in vitro. J Neurosci 1994; 14: 2674–2686.
Martin DP, Schmidt RE, DiStefano PS, Lowry OH, Carter JG, Johnson EMJ . Inhibitors of protein synthesis and RNA synthesis prevent neuronal death caused by nerve growth factor deprivation. J Cell Biol 1988; 106: 829–844.
Du Y, Bales KR, Dodel RC, Hamilton-Byrd E, Horn JW, Czilli DL et al. Activation of a caspase 3-related cysteine protease is required for glutamate-mediated apoptosis of cultured cerebellar granule neurons. Proc Natl Acad Sci USA 1997; 94: 11657–11662.
Wood KA, Dipasquale B, Youle RJ . In situ labelling of granule cells for apoptosis-associated DNA fragmentation reveals different mechanisms of cell loss in developing cerebellum. Neuron 1993; 11: 621–632.
Bobba A, Atlante A, Giannattasio S, Sgaramella G, Calissano P, Marra E . Early release and subsequent caspase-mediated degradation of cytochrome c in apoptotic cerebellar granule cells. FEBS Lett 1999; 457: 126–130.
Miller TM, Moulder KL, Knudson CM, Creedon DJ, Deshmukh M, Korsmeyer SJ et al. Bax deletion further orders the cell death pathway in cerebellar granule cells and suggests a caspase-independent pathway to cell death. J Cell Biol 1997; 139: 205–217.
Ham J, Babij C, Whitfield J, Pfarr CM, Lallemand D, Yaniv M et al. A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron 1995; 14: 927–939.
Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 1997; 91: 231–241.
Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999; 96: 857–868.
Gilley J, Coffer PJ, Ham J . FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol 2003; 162: 613–622.
Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J . Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron 2001; 29: 629–643.
Putcha GV, Harris CA, Moulder KL, Easton RM, Thompson CB, Johnson Jr EM . Intrinsic and extrinsic pathway signaling during neuronal apoptosis: lessons from the analysis of mutant mice. J Cell Biol 2002; 157: 441–453.
Linseman DA, Butts BD, Precht TA, Phelps RA, Le SS, Laessig TA et al. Glycogen synthase kinase-3beta phosphorylates Bax and promotes its mitochondrial localization during neuronal apoptosis. J Neurosci 2004; 24: 9993–10002.
Knudson CM, Tung KS, Tourtellotte WG, Brown GA, Korsmeyer SJ . Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science 1995; 270: 96–99.
Towers E, Gilley J, Randall R, Hughes R, Kristiansen M, Ham J . The proapoptotic dp5 gene is a direct target of the MLK-JNK-c-Jun pathway in sympathetic neurons. Nucleic Acids Res 2009; 37: 3044–3060.
Arce V, Garces A, de Bovis B, Filippi P, Henderson C, Pettmann B et al. Cardiotrophin-1 requires LIFRbeta to promote survival of mouse motoneurons purified by a novel technique. J Neurosci Res 1999; 55: 119–126.
Lutfalla G, Uze G . Performing quantitative reverse-transcribed polymerase chain reaction experiments. Methods Enzymol 2006; 410: 386–400.
Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME . Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 1995; 270: 1326–1331.
Aalto AP, Sarin LP, van Dijk AA, Saarma M, Poranen MM, Arumae U et al. Large-scale production of dsRNA and siRNA pools for RNA interference utilizing bacteriophage phi6 RNA-dependent RNA polymerase. RNA 2007; 13: 422–429.
Swainson L, Mongellaz C, Adjali O, Vicente R, Taylor N . Lentiviral transduction of immune cells. Methods Mol Biol 2008; 415: 301–320.
Acknowledgements
This work was supported by the Centre National de la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM), the Université Montpellier 2, the Fondation pour la Recherche Médicale (FRM), the Association pour la Recherche sur le Cancer (ARC) and the Wellcome Trust. We are grateful to the staff of the animal facilities of the IGMM for the breeding of mice and services in histology, the RIO imaging platform for technical assistance, the recombinant protein production facility of the IFR122 of Montpellier for the production and the purification of the Trim17 antibody and the vectorology platform of the IFR3 of Montpellier for technical assistance in the production of lentiviral particles. We thank Dr. I Allemand for the gift of Bax-deficient mice, Dr. O Coux for in vitro ubiquitination reagents and Dr. G Sczakiel for help in the definition of the siRNA no. 1 target sequence. We are grateful to Dr. J-C Martinou for critical reading of the article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Lassot, I., Robbins, I., Kristiansen, M. et al. Trim17, a novel E3 ubiquitin-ligase, initiates neuronal apoptosis. Cell Death Differ 17, 1928–1941 (2010). https://doi.org/10.1038/cdd.2010.73
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2010.73
Keywords
This article is cited by
-
The E3 ubiquitin ligase TRIM17 promotes gastric cancer survival and progression via controlling BAX stability and antagonizing apoptosis
Cell Death & Differentiation (2023)
-
Tripartite Motif Protein Family in Central Nervous System Diseases
Cellular and Molecular Neurobiology (2023)
-
Gene losses may contribute to subterranean adaptations in naked mole-rat and blind mole-rat
BMC Biology (2022)
-
TRIM17 and TRIM28 antagonistically regulate the ubiquitination and anti-apoptotic activity of BCL2A1
Cell Death & Differentiation (2019)
-
TRIM22-Mediated Apoptosis is Associated with Bak Oligomerization in Monocytes
Scientific Reports (2017)