Abstract
p73 and p63, the two ancestral members of the p53 family, are involved in neurogenesis, epithelial stem cell maintenance and quality control of female germ cells. The highly conserved oligomerization domain (OD) of tumor suppressor p53 is essential for its biological functions, and its structure was believed to be the prototype for all three proteins. However, we report that the ODs of p73 and p63 differ from the OD of p53 by containing an additional α-helix that is not present in the structure of the p53 OD. Deletion of this helix causes a dissociation of the OD into dimers; it also causes conformational instability and reduces the transcriptional activity of p73. Moreover, we show that ODs of p73 and p63 strongly interact and that a large number of different heterotetramers are supported by the additional helix. Detailed analysis shows that the heterotetramer consisting of two homodimers is thermodynamically more stable than the two homotetramers. No heterooligomerization between p53 and the p73/p63 subfamily was observed, supporting the notion of functional orthogonality within the p53 family.
Similar content being viewed by others
Main
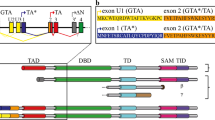
p53, a well-known tumor suppressor that is mutated in more than 50% of all human tumors, induces genes leading either to cell-cycle arrest or to apoptosis. The discovery of two proteins with a high sequence identity to p53, called p63 and p73, has sparked speculations that tumor suppression is carried out by the combined action of several members of this protein family, for example, by direct interaction through heterooligomerization. Knockout mouse studies with p63 and p73, the two ancestral members,1 have shown functional roles distinct from p53: p63 is essential for maintaining epithelial stem cells2, 3 and for protecting the genomic stability of oocytes,4 whereas p73 is involved in neurogenesis, sensory pathways and homeostatic control.5 p73 is further known as an important inducer of apoptosis in response to DNA damage.6 Although only few mutations of p63 and p73 have been found in human tumors so far, overexpression of p63 is often observed in squamous cell carcinoma,7, 8, 9, 10 which has been shown to suppress p73-dependent apoptosis.11 Among all p53 family members, including those from invertebrate species, the DNA binding domain is the most conserved domain,12, 13, 14, 15 followed by the oligomerization domain (OD), which is indispensable for the biological function of all p53 protein family members.16, 17, 18 It is a structural domain that forms a tetramer, and mutations within the OD that inhibit tetramerization of p53 result in greatly reduced transcriptional activity.19 In addition, several protein–protein interactions and posttranslational modifications require the tetrameric state as well,16 and mutations in the OD of p53 that prevent oligomerization have been identified in human cancers.20, 21 Owing to its functional importance, the OD of p53 has been the target of several structure determination projects.22, 23 The p53 tetramer consists of a dimer of dimers, with each monomer contributing one β-strand and one α-helix. Dimers are assembled by the formation of an intermolecular antiparallel β-sheet, which is stabilized by hydrophobic interactions with two helices that also arrange in an antiparallel orientation. The tetramer is created by hydrophobic interactions between the helices of both dimers. This arrangement was so far considered to be the prototype for all members of the p53 family. Figure 1 shows a comparison of the OD sequences of all three human family members, showing that significant sequence identity exists in the region covering the β-strand and the α-helix. C-terminal to the helix, however, sequences diverge into the p53 subgroup and into the p73/p63 subgroup. In p53, an unstructured, protease-sensitive stretch of 38 amino acids is found. This C-terminal domain is the target of many posttranslational modifications and was shown to be important for the regulation of DNA-binding activity.24 In the α-splice forms of p63 and p73, a long stretch of amino acids (119 in p63 and 109 in p73) connects the OD to a sterile alpha motif (SAM) domain, which is followed by a transcriptional inhibitory domain.12, 25 Several invertebrate p53 proteins also contain long C-terminal stretches, however, with low sequence identity to the mammalian family members. Surprisingly, in the p53 forms of the two important model organisms, Caenorhabditis elegans (CEP-1) and Drosophila melanogaster (Dmp53), this low sequence identity includes the otherwise highly conserved OD. Structure determination of the C-termini of CEP-1 and Dmp53 showed that both proteins contain an OD, however, with architectures different from that of p53.26 In particular, we showed that CEP-1 is the first known dimeric member of this protein family and that its OD is structurally coupled to a SAM domain. Dmp53 showed yet another architecture. Its tetrameric state is stabilized by an additional N-terminal β-strand and an additional C-terminal α-helix, thus doubling all secondary structure elements present in the p53 OD.26 These structure determinations of the ODs of invertebrate forms have cast the first doubt on the universal structural role of the p53 OD. These results, combined with the high sequence conservation of ∼20 amino acids C-terminal to the OD of p63 and p73 (Figure 1), and the fact that p63 inhibits the transcriptional activity of p73 and directly binds to it in head and neck squamous cell carcinoma,11 prompted us to reinvestigate the structure of the OD of these two proteins.
Sequence alignment of the oligomerization domains of members of the human p53 family. All isoforms of members of the human p53 family contain an OD with high sequence identity to the OD of p53 (40% with p63 and 43% with p73). The sequence identity between p63 and p73 in the region of the canonical OD reaches 60%. Residues that are conserved among all members of the family are shown in green, whereas residues that are only conserved between p63 and p73 are colored blue. The secondary structure elements known from p53 OD are indicated on top of the figure. The p53 sequence diverges from other sequences C-terminal from the helix. p63 and p73 show high sequence identity over an additional stretch of ∼20 amino acids (383–398 in p73, 390–410 in p63), after which sequences even among different isoforms start to differ. Several p63 and p73 isoforms encompass sequences and domains C-terminal to the region shown in this figure
Results
The tetrameric state of p73 is stabilized by an additional helix
To investigate whether direct interaction between p63 and p73 occurs through their highly homologous ODs, we intended to perform nuclear magnetic resonance (NMR)-based titration experiments. Surprisingly, the spectrum of the p73 OD (consisting of the canonical β-strand and the α-helix, amino acids 351–381), which is shown in Figure 2a, showed a more than doubling of the number of expected resonances that can be reduced to the anticipated 31 peaks by lowering the temperature to 2°C (Figure 2b), suggesting the existence of different oligomeric states. This interpretation was confirmed by further investigation by analytical ultracentrifugation that showed an equilibrium between dimers and tetramers (Supplementary Figure 1). Additional NMR experiments of this dimer–tetramer mixture suggested the existence of three different species (Supplementary Figure 2). As mentioned in the Introduction, structure determination of the OD of Dmp53 showed the existence of one additional N-terminal β-strand and one additional C-terminal α-helix compared with that of p53 OD.26 Removal of either noncanonical structural element led to a conformational instability of the Dmp53 OD, thus demonstrating that these additional structural elements are essential for the stabilization of the structure and the oligomeric state of this domain. Interestingly, secondary structure prediction of the p73 sequence suggested the existence of an additional helix C-terminal to its canonical OD. To verify its existence and to investigate its influence on the structure and on the dimer–tetramer equilibrium, we recorded NMR spectra of an extended p73 OD ranging from 351 to 398 amino acids (construct subsequently referred to as tetramerization domain, TD). As evident from the spectrum shown in Figure 2c, only one set of resonances was detectable even at 30°C. Moreover, many resonances showed chemical shift differences compared with the shorter construct, suggesting that the additional amino acids interact with the core of the TD (Figure 2d). The tetrameric state of this species was confirmed both by analytical ultracentrifugation (Supplementary Figure 1 and Supplementary Table 1) and by measurement of the rotational correlation time by NMR spectroscopy (τc=12 ns). Analysis of the secondary structure content from chemical shift data confirmed the existence of a second helix at the C-terminus of the construct (amino acids 383–396) (Supplementary Figure 3). To investigate whether additional secondary structure elements exist either N-terminal to the β-strand or C-terminal to the second helix of the TD, we recorded NMR spectra of extended constructs. These spectra, however, did not show any resonance changes in TD, whereas the resonances of the added amino acids showed random coil characteristics, suggesting that no additional secondary structure elements were present.
NMR spectra of C-terminally extended p73 oligomerization domains. (a) The [15N, 1H]-TROSY spectrum of p73 OD (351–381) encompassing the region of the β-strand and the α-helix, measured at 30°C. The spectrum shows more than twice the number of expected resonances. (b) Spectrum of the same sample as in a, measured at 2°C. Only a single set of peaks is observed. (c) Extending the construct toward the C-terminus (351–398) resulted in a single set of resonances even at 30°C. (d) Comparison of [15N, 1H]-TROSY spectra of both p73 OD constructs (351–381 in red, 351–398 in blue) at 2°C. In addition to a larger number of peaks, the spectrum of the longer p73 construct shows widespread differences in chemical shifts relative to the short form, suggesting that the added amino acids interact with the core of the oligomerization domain
Structure determination of TD confirmed the existence of the helix, as well as its interaction with the TD core domain (Figure 3). The core domain consisting of the β-strand and the first α-helix shows a tetrameric arrangement similar to that of the p53 OD. The transition from the last turn of helix I with a 310-helical geometry to helix II occurs over Pro 382. Helix II of monomer A (as defined in Figure 3) reaches across the tetrameric interface and forms hydrophobic interactions with helix I of monomer C encompassing Leu 377, Leu 380, Val 381, Leu 385, Val 386 and Tyr 389. Apart from these hydrophobic interactions, hydrogen bonds between Gln 393 (monomer A) and Glu 373 (monomer C) are possible. Glu 373 (monomer C) also forms a salt bridge with Arg 390 (monomer A). This salt bridge is further supported by heteronuclear nuclear Overhauser effect (NOE) experiments (Supplementary Figure 4) that showed that the ɛ-NH group of Arg 390 is conformationally restricted.
Structure of the p73 tetramerization domain (TD). (a) Sequence and secondary structure of p73 TD and p63 TD. (b) Structure of the TD of p73. The four individual monomers are labeled as A, B, C and D. The color of the letters corresponds to the color of the monomers. Dotted lines in the second helices indicate hydrogen bonds that were determined by measuring direct coupling between amide protons and carbonyl carbons. (c) A view of the structure of the TD of p73 along another C2 axis of the molecule
Mutation of Arg 390, Gln 393 and Tyr 389 leads to conformational instability
To investigate the importance of Arg 390 and Gln 393 in the stabilization of the tetrameric interface, we mutated them to methionine and alanine, respectively. Although the single-point mutations, Arg390Met and Gln393Ala, did not affect the oligomeric state as investigated by NMR, the double mutation, Arg390Met/Gln393Ala, gave rise to more than two sets of peaks at 30°C (Supplementary Figure 5) and a single set at 2°C, indicating the presence of a dimer–tetramer equilibrium. In addition to Arg 390 and Gln 393, the structure of TD suggested that Tyr 389 is of central importance for stabilizing hydrophobic contacts between monomers. The mutant Tyr389Ala showed more than two sets of resonances at 30°C (Supplementary Figure 5) and one set at 2°C, thus yielding a similar result as the double mutation Arg390Met/Gln393Ala or when helix II is deleted. The importance of Tyr 389 for the stability of the tetramer is further apparent from its strict conservation in all p73 sequences (Figure 1 and Supplementary Figure 6).
Interestingly, this tyrosine is also conserved in p63. Furthermore, Arg 390 and Gln 393 are conserved in all p63 isoforms as well, suggesting that the p63 OD is also stabilized by an additional helix. Assignment of the NMR resonances of TD and the chemical shift-based secondary structure analysis indeed confirmed the existence of a second helix in p63 (Figure 3a).
The C-terminal helix of p73 is necessary for high transcriptional activity
Several studies have shown that the tetrameric state is important for the transcriptional activity of p53. Furthermore, mutations in the OD of p53 have been found in cancer patients, providing a further link between tetrameric state and activity. To test whether the additional helix present in the TD of p73 not only influences the conformational stability of TD but also the transcriptional activity of the entire protein, we deleted the second helix in the shortest p73 isoform, TAp73δ,27 which ends a few amino acids C-terminal to the second helix. Transactivation assays in SAOS-2 cells on the p21 and on the bax promoter showed a high transcriptional activity of the β-isoform and a reduced activity of the δ-isoform on both promoters as known from earlier investigations27 (Figure 4). Deletion of the last helix led to a strong reduction in transcriptional activity on the p21 promoter and to a smaller reduction on the bax promoter. Mutation of the critical Tyr 389 to alanine reduced transcriptional activities on both promoters as well. These data provide a link between the second helix and the transcriptional activity of p73 and suggest that the core OD, as known from the crystal and NMR structure of the p53 OD, is not sufficient to guarantee a high transcriptional activity. Our results, however, also suggest that the effect of the second helix depends on the nature of the promoter.
Influence of the second helix of TD on the transcriptional activity of p73. (a) The transcriptional activity of the β- and δ-isoform, as well as of the TAp73δ isoform mutants lacking either the second helix or bearing the Y389A point mutation, was measured both on the p21 promoter (blue) and on the bax promoter (red) in SAOS-2 cells. The western blots shown in (b) were used to normalize transcriptional activity according to the intracellular p73 concentration. GAPDH was used as an internal standard in western blot analysis, and the intracellular p73 concentration was corrected for transfection efficiency. Each measurement and western blot was carried out in triplicate. Error bars indicate the maximum deviation from the mean value
p73 TD and p63 TD form several heterotetramers
Earlier, the high sequence conservation between the ODs of p53, p63 and p73 had already sparked speculation about the potential formation of heterotetramers between all three proteins. However, so far, only a very weak interaction between the ODs of p63 and p73, and no interaction of either of the ODs of these proteins with p53, has been reported.28 In contrast to these biochemical studies with purified ODs, interaction between p63 and p73 isoforms has been reported in mammalian cells.11 As the TDs of p63 and p73 show identical structural topology and exhibit high sequence conservation, we investigated their potential heterooligomerization by recording NMR spectra of mixtures of the TDs of p73 (15N-labeled) and p63 (unlabeled) in equal amounts. The [15N, 1H]-TROSY (transverse relaxation-optimized spectroscopy) spectrum recorded after denaturation of the mixture in 6 M Gdn-HCl and the subsequent refolding showed many additional resonances consistent with the presence of different heterooligomeric states (Supplementary Figure 7). To analyze the heterooligomerization in a more quantitative manner, we performed the same experiment with p73 TD selectively 15N-labeled on lysines. The resulting [15N, 1H]-TROSY spectrum (Figure 5a) showed eight different peaks for Lys 372, which is in agreement with the number of possible heterooligomers (Supplementary Figure 7), indicating that p73 and p63 can form heterooligomers with all statistically possible stoichiometry. The tetrameric state of the heterooligomers was also confirmed by size-exclusion chromatography (data not shown). To test for the possibility that heterooligomerization occurs only after denaturation and subsequent refolding, we mixed the folded TDs of p73 (15N-labeled on lysines) and p63 in equal amounts at 37°C. After 5 h, additional peaks corresponding to the resonances observed in the spectrum in Figure 5a appeared, showing that dimer and monomer exchange between p63 and p73 tetramers occurs even when both domains are folded (Figure 5b and Supplementary Figure 8). Surprisingly, the most prominent peak in the spectrum after 5 h is peak 3, which belongs to a heterooligomeric species. Very similar results were obtained by mixing 15N-lysine-labeled p63 TD with unlabeled p73 TD (Figure 5c and d).
Heterooligomerization between the TDs of p73 and p63 and coexpression in epithelial tissue. (a) Section of an [15N, 1H]-TROSY spectrum of the p73 TD selectively labeled on lysines, showing Lys 372. The sample was prepared by mixing labeled p73 TD and unlabeled p63 TD, followed by denaturation with Gdn-HCl and subsequent refolding. A total of eight different resonances can be detected. The peak position of Lys 372 in the homotetrameric p73 TD is shown in blue. (b) Section of a spectrum obtained by mixing the TDs of p73 (lysine labeled) and p63 (unlabeled) without denaturation. The small insert shows the spectrum taken 1 h after mixing, which demonstrates that at this time only one heterooligomeric species has appeared. After 5 h, all eight resonances were detectable. (c) The [15N, 1H]-TROSY spectrum of 15N-lysine-labeled p63 TD, mixed with unlabeled p73 TD without denaturation, showing the resonances of Lys 377 after 1 hour at 37°C. The peak corresponding to the homotetrameric p63 TD is shown in blue. (d) The same experiment as depicted in c, showing the spectrum after an incubation time of 5 hours at 37°C. (e) Populations of different heterotetramers based on the spectra shown in b and d are depicted in red. The theoretical statistical distribution is shown in black. Heterodimer-containing tetramers are populated lower than expected from statistics. The predominant heterotetramer (represented by peaks 3 and 9) consists of two different homodimers and its population (∼44%) exceeds the ratio of 2 : 1 relative to that of homotetramers (represented by peaks 2 and 11), which is expected from a statistical distribution. Error bars represent standard deviations in peak integration estimated from repeated peak volume determinations. (f) Mouse skin section stained with the monoclonal anti-p63 antibody 4A4. (g) A similar skin section stained with the monoclonal anti-p73 antibody 3A3. These sections show that both proteins are coexpressed in the basal cells of epithelial tissue
A p73/p63 heterotetramer is more stable than homotetramers
To obtain a more quantitative analysis of the populations of various heterospecies, we assigned the peaks in the spectra shown in Figure 5b and d on the basis of their relative peak volumes and time of appearance. Peak 3 (Figure 5b) and peak 9 (Figure 5d) represent a heterotetramer consisting of two homodimers, as they are the first peaks to arise after mixing. The formation of heterodimer-containing tetramers would be associated with the simultaneous appearance of more peaks. The remaining peaks in both spectra were assigned on the basis of considerations described in experimental procedures. On the basis of this peak assignment, it was possible to calculate the populations of heterotetramers (Figure 5e). A comparison of these populations with expected populations from a purely statistical distribution (assuming no difference in thermodynamic stability between the different heterooligomeric and homooligomeric species) showed significant deviations. Interestingly, heterodimer-containing tetramers were populated lower than expected. In contrast, the heterotetramer that consists of two homodimers is approximately four times more highly populated than homotetramers, and therefore more stable.
To further investigate the importance of the additional C-terminal helix, we performed experiments with the ODs of p63 and p73 (the TD lacking the second helix). Mixing of 15N-labeled p73 OD and unlabeled p63 OD showed that in this mixture of different homodimeric and homotetrameric species that exist without the stabilizing effect of the C-terminal helix (Figure 2a), only one additional heterooligomer is formed (Supplementary Figure 9), thus showing that the additional C-terminal helix has a strong influence on the formation of heterooligomeric species.
The TDs of p73 and p63 do not interact with p53 OD
p53 and p73 (or p63) are coexpressed in many tissues. We, therefore, wanted to investigate the potential of both proteins to form heterooligomers as well. However, mixing of either p73 TD or p63 TD with p53 OD did not result in the formation of any heterooligomers on the basis of the observation of only one set of peaks in [15N, 1H]-TROSY spectra (Supplementary Figure 10).
In contrast, we observed an additional set of peaks from mixtures of p73 OD (i.e., TD lacking helix II) with p53 after denaturation and refolding, consistent with the formation of heterooligomers (Figure 6). These heterooligomers seemed to be less stable than homooligomers, because their peak intensity was strongly reduced over a period of 26 h (Supplementary Figure 10). Mixing of the folded p73 OD and p53 OD, however, resulted in the same low amount of heterooligomer. Interestingly, the p63 OD did not show any interaction with p53 even after denaturation.
Heterooligomerization of p73 OD with p53 OD. (a) The [15N, 1H]-TROSY spectrum of uniformly 15N-labeled p73 OD mixed with an equimolar amount of p53 OD. Before performing NMR measurements, the mixture was denatured with 6M Gdn-HCl and refolded. The spectrum in red, showing one set of additional strong resonances compared with the spectrum of 15N-labeled p73 OD (in blue) was taken immediately after refolding. (b) Spectrum of the same sample as in a measured after incubation for 1 day at 30°C and for an additional 3 days at 37°C. Arrows exemplarily show three remaining resonances of the p73 OD/p53 OD heterotetramer. The same spectrum was obtained after mixing uniformly 15N-labeled p73 OD and p53 OD without denaturation and an incubation time of 3 days at 37°C. (c) The p53 OD/p73 OD heterotetramer that was formed after refolding is mainly converted into three different p73 OD conformers and the p53 OD tetramer
Discussion
The experiments described above indicate that the ODs of p73 and p63 contain an additional helix not present in p53. In the case of p73, this additional helix stabilizes the tetrameric state, which results in a decrease in the dissociation constant kD from 50 μM (OD without the helix, assuming a simple dimer–tetramer equilibrium) to 3 μM. This value is still too high to ensure that p73 is predominantly tetrameric in vivo; however, the equilibrium might be further affected by other domains in the full-length protein, such as the DBD and the SAM domain. In addition, the interaction with other proteins may influence the oligomeric state, as it was shown for p53.29, 30
Taken together, the structures of the ODs of Cep-1 and Dmp53, the structure of the p73 TD as shown in this study and the secondary structure assignment of the p63 TD show that the OD of p53 is an exception within this protein family; it is the only one that does not contain additional domains (Cep-1) or secondary structure elements (p63, p73, Dmp53).
Very recently, Nyman et al.31 identified a second transactivation domain in the C terminus of p73. They showed that this new transactivation domain preferentially regulates genes involved in cell-cycle arrest. They mapped the location of this transactivation domain to amino acids 381–399, which almost exactly coincides with the second helix that we have identified in the TD (amino acids 383–395). The finding that this helix is important for the preferential activation of cell-cycle arrest versus apoptosis, combined with the importance of this helix for the oligomeric state and the conformational stability of p73, leads to the intriguing speculation that the oligomeric state has a role in the decision to induce either cell cycle arrest or apoptosis. Although tetramerization – and therefore the second helix – might be necessary to induce cell-cycle arrest, apoptosis could be regulated by dimeric forms or forms with a different geometric arrangement that do not rely on the second helix for stabilization. Our own cell culture experiments have suggested that deletion of the second helix has a more significant effect on the transcriptional activity of p73 on the cell-cycle arrest p21 promoter than on the apoptosis bax promoter. Nyman et al. further showed that Ser 388 becomes phosphorylated by protein kinase C. In our structure, Ser 388 is not involved in the tetramerization interface but solvent exposed and accessible for kinases. This modification does not influence the structure of TD directly, but most likely provides a binding site for additional factors that might influence the oligomeric state of the protein. Different posttranslational modifications of TD that attract various proteins might therefore provide a basis for forming distinguished oligomeric states or conformations of the entire protein. Proteins that interact with members of the p53 protein family and that affect the oligomeric state have been identified.29, 30 Furthermore, it is intriguing to speculate that the second helix of TD that is present in p63 and in p73 as a structural element has become a regulatory element in p53, in which it might form in the C-terminal regulatory domain (367–393). The transition from the unfolded state to a helical one, which stabilizes the tetrameric conformation, might be triggered by a specific pattern of posttranslational modification or by interaction with other binding partners.
The additional helix within the TDs of p63 and p73 also influences the interaction among the individual members of this protein family. Although it inhibits even transient interactions between p73 (or p63) and p53, it enhances the interaction between p63 and p73. Despite mouse knockout studies showing disparate functions for p63 and p73, recent reports suggest that the two genes are coexpressed in various cell lines and tissues. Immunoprecipitation experiments in JHU 029 cells, a cell line derived from head and neck squamous cell carcinoma (HNSCC), have shown that the majority of TAp73β is physically bound to ΔNp63α.11 In these cells, the high expression of ΔNp63α inhibits p73-dependent apoptosis. RNAi-based knockdown experiments of p63 in these cell lines led to the expression of NOXA and PUMA in a p73-dependent but p53-independent manner, thus showing that the high expression of ΔNp63α constitutes an antiapoptotic and prosurvival signal. Our structural investigations reported here show that p73 and p63 preferentially form heterotetramers, consisting of two homodimers, thus revealing for the first time the structural basis for the interaction of p73 and p63 in HNSCC cells.
The interaction between p63 and p73 is most likely not only relevant for cancer cells but also has a role in the basal compartment of epithelial tissue. Staining of mouse skin sections showed that both proteins are highly expressed in these basal cells (Figure 5c and d). The relative expression level of p63:p73 is roughly 10 : 1, suggesting that a large percentage of p73 is bound in heterooligomeric complexes as well. The exact function of heterotetramers formed by ΔNp63α and ΔNp73α in basal cells, however, is currently not understood. The data presented here suggest that, in the future, p63 and p73 might have to be considered as partners that interact with each other and, at least in HNSCC cells, have opposing effects. On the other hand, active participation in tumor suppression on the basis of direct interaction with p53 does not seem to occur.
Materials and Methods
Protein expression and purification
The following constructs were used in this study: human p73 TD (351–398), p73 OD (351–381 or 351–384), mouse p63 TD (358–416), p63 OD (358–391) and mouse p53 OD (320–356). The amino acids were numbered as defined for TA isoforms (GenBank accession numbers: CAA72219 for human p73, AAC62635 for human p63, AAC62641 for mouse p63 (deducted 39), NP_001119584 for human p53 and NP_035770 for mouse p53). The TDs of mouse and human p63 are identical in sequence, except for Met 414 (Ile in human), which is located in the unstructured region C-terminal to helix II. The ODs of mouse and human p53 differ only in amino acids Lys 328 (Gln 331 in human), Lys 333 (Glu 336 human) and C-terminal of Ala 350. Although the latter segment is not structured in the human p53 OD, the amino acids Gln 331 and Glu 336 are solvent exposed. For that reason, these mouse constructs can also be used in heterooligomerization studies with human p73 constructs.
All p73, p63 and p53 constructs were cloned into the plasmid pBH4 (gift from Wendell Lim laboratory, San Francisco, CA, USA) containing an N-terminal tobacco etch virus protease (TEV)-cleavable His6 tag and were expressed in the E. coli strain T7-Express (NEB #C2566H) or DL39 (for Val-labeled samples). Cells were grown in a 2XYT medium to an OD600 of 0.6. For the expression of isotopically labeled proteins, cells were pelleted and carefully resuspended in half of the original volume of M9 media supplemented with [u-13C6]-D-glucose, [15N]-ammonium chloride or either [15N]-Valine or [15N]-lysine. Proteins were purified by Ni-affinity chromatography and dialyzed overnight during TEV-mediated cleavage. TEV (with His6-tag) and uncleaved proteins were removed in a second Ni-chelation affinity chromatography step. The flow through was collected and further purified by gel filtration chromatography using a Superdex 75 column (GE Healthcare, München, Germany). The protein solution was concentrated using Amicon Ultra-4 Ultracells (Millipore, Schwalbach, Germany). Protein samples were stored at −80°C and measured in buffer containing 20 mM sodium phosphate (pH 7.0), 100 mM sodium chloride, 0.5 mM EDTA or 50 mM arginine/glutamate (pH 6.8). For NMR spectroscopy, [u-15N]- and [u-15N,13C]-labeled protein samples were used at concentrations of 2.5 mM. For the measurement of 13C-edited nuclear Overhauser effect spectroscopy (NOESY) and [1H, 1H]-NOESY spectra, samples were exchanged to D2O by lyophilization and pD was adjusted with NaOD.
For the measurement of intersubunit NOEs and h3J couplings, [u-13C]- and [u-15N]-labeled proteins were mixed in equal amounts, denatured in guanidine hydrochloride and stirred for 1 h at room temperature. The denatured protein mixture was slowly dripped into 160 ml of 20 mM sodium phosphate (pH 7.0), 100 mM sodium chloride, 0.5 mM EDTA, concentrated with an Amicon ultrafiltration membrane and purified by gel filtration chromatography using a Superdex 75 column.
NMR spectroscopy
All NMR experiments were performed using Bruker (Rheinstetten, Germany) Avance spectrometers equipped with 1H-triple resonance, z-gradient cryogenic probes at proton frequencies of 950, 900, 800, 700 or 600 MHz, or on a Bruker 500 MHz spectrometer equipped with a room-temperature triple resonance, x,y,z-gradient probe. All experiments were measured at 303 K unless otherwise stated. DSS (4,4-dimethyl-4-silapentane-1-sulphonate) was used as an internal chemical shift reference. Spectra were processed with Bruker Topspin 2.1 or NMRPipe (NIH, Bethesda, MD, USA) and were analyzed using UCSF SPARKY 3.114 (San Francisco, CA, USA).32 Protein backbone resonances were assigned with TROSY-based HNCACB, HNCOCA and HNCO experiments.33, 34 Side-chain resonances were assigned with H(CCCO)NH, (H)C(CCO)NH, 3D TOCSY-[15N,1H]-TROSY and 13C-separated 3D NOESY experiments. The prochiral methyl groups of valine and leucine with a CT-[13C,1H]-HSQC were assigned by 10% nonrandom 13C protein labeling. The side-chain amide protons of Asn and Gln residues were stereospecifically assigned using an H2NCO-E.COSY experiment. Aromatic side chains were assigned using 2D (HB)CB(CGCC-TOCSY)Har experiments.35 With the exception of the δ-proton and carbon resonances of Lys 372 and Arg 398, all other backbone and side-chain resonances were assigned.
Dihedral angles were obtained with TALOS using the chemical shifts of H, N, Cα, Cβ and CO.36 Distance constraints were extracted from 3D NOESY-[13C,1H]-HSQC (in H2O and D2O), 3D NOESY-[15N,1H]-TROSY (in H2O) and 2D-NOESY (in D2O) experiments with a mixing time (τM) of 50 ms. Intersubunit distance constraints were obtained with 4D-CT-J-Resolved 13C-separated NOESY (in D2O, τM=100 ms),37 with 3D 15N-edited/13C-separated NOESY (τM=140 ms; in H2O) and with 3D 15N/13C-separated NOESY (τM=110 ms, in H2O) experiments. Intermolecular H bonds in the β-strand and intramolecular H bonds in α-helices were detected with a 3D-[15N,1H]-TROSY long-range HNCO (Supplementary Table 3). In addition, amide protons that were still present after 5 days of H/D exchange were assumed to participate hydrogen bonds. Residual dipolar couplings were determined by weakly aligning the protein with the filamentous phage Pf1 (Profos, Regensburg, Germany). RDCs were measured with a Cα-coupled HA(CACO)NH and an F2-IPAP-CT-[13C,1H]-HSQC for 1D(Hα-Cα), a [15N,1H]-TROSY/anti-TROSY for 1D(HN-N), an HN(α/β-NC'-J)-TROSY IPAP for 2D(HN-C)/1D(N-C) and a Cα-coupled HNCO for 1D(C-Cα). The initial estimate for the alignment tensor was determined with the extended histogram method.38 For scaling the magnitude Da of the alignment tensor to the appropriate internuclear vector, an average order parameter S of 0.94 was assumed for the HN–N pairs. In addition, order parameters and internuclear distances for atom pairs were set as described in reference.39
HetNOEs were measured with an 15N-[1H]-NOE-TROSY experiment. The errors result from peak volume fitting inaccuracies.
The global rotational correlation time of p73 TD was determined using the [15N,1H]-TRACT experiment.
To determine the populations of diverse p73/p63 heterotetramers, peak volumes were extracted (using Sparky, ellipse integration method, measured thrice manually). To join the information from the two spectra, the relative peak volumes of peak 3 (Figure 5b) and peak 9 (Figure 5d) were used for normalization. The peaks corresponding to those heterotetramers that contain three labeled p63 or three labeled p73 molecules were assigned using their characteristic peak pattern (one peak for each labeled molecule inside the tetramer because of lack of symmetry; peaks resulting from one heterotetramer exhibit identical relative peak volumes). Peak volumes were divided by the number of molecules that they represent and were finally normalized.
Structure calculation
Structure calculations were carried out using Aria 2.2/CNS40 (Institut Pasteur, Paris, France), with modified protocols that imposed a D2 symmetry throughout every stage of calculation. The procedure used to identify intermonomer and interdimer NOEs has been described before.26 The structure is based on 908 intrasubunit NOEs, 340 intersubunit NOEs within a dimer and 242 intersubunit NOEs across the tetrameric interface. A total of 20 structures were calculated in 8 iterations, and 100 structures were calculated in the last iteration. To deal with NOE ambiguities arising from the tetrameric state of the protein, 144 000 cooling steps were performed during simulated annealing. RDCs were introduced in the eighth iteration. RDC force constants (kH−N=10, kCA−HA=0.8, kH−C=5, kN−C=10, kC−CA=10; in kcal Hz−2) were adjusted so that the experimental error (ΔH−N=0.19, ΔCA−HA=0.98, ΔH−C=0.26, ΔN−C=0.05, ΔC−CA=0.15; in Hz) is lower than the root-mean square deviation of RDCs. After multiple iterations of peak inspections and structure calculations, 100 structures were calculated and the best 20 structures were used for water refinements and analysis. All distance constraint violations were smaller than 0.2 Å. The structural statistics can be found in Supplementary Table 2.
Analytical ultracentrifugation
Ultracentrifugation experiments were carried out at a temperature of 8°C, in 20 mM of either Tris/HCl or Bis-Tris propane, each at pH 7.8 and containing 150 mM NaCl. Sedimentation of particles was monitored using interference optics in a Beckman (High Wycombe, UK) XL-I Analytical Ultracentrifuge equipped with a Ti-50 rotor. Sedimentation velocity experiments were performed at monomer concentrations of 50 μM in two-sector cells, using a rotor speed of 50 000 r.p.m. Radial scans were collected at 30-s intervals. Data were analyzed using SEDFIT (NIH) to calculate c(s) distributions. The software package SEDNTERP (Thousand Oaks, CA, USA) was used to normalize the obtained sedimentation coefficient values to the corresponding values in water at 20°C. Sedimentation equilibrium experiments were performed at 8°C at rotor speeds of 17 000, 22 000 and 31 000 r.p.m., and at protein concentrations of 10, 25 and 50 μM. Working on the assumption that monomers do not exist in solution, the dissociation constant was calculated from a global fitting of all data to a monomer–dimer equilibrium model, using ULTRASPIN (MRC Centre for Protein Engineering, Cambridge, UK).
Transactivation assays
p73 constructs for cell culture experiments were cloned into the pCDNA3.1 vector containing an N-terminal myc tag. Inserts similar to p73 isoforms β and δ were generated from a p73-α template: TAp73β (1–494), TAp73δ (1–399) and TAp73δΔH2 (1–381). All transactivation experiments were performed in SAOS-2 cells using the Promega (Mannheim, Germany) Dual-Glo Luciferase reporter assay. For transfection assays, a construct with either a single copy of p21 or the bax promoter obtained from human genomic DNA was cloned into the pGL3 vector. Cells were obtained from ATCC (Manassas, VA, USA) and were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum at 37°C under an atmosphere of 5% CO2. Cells were transfected with 133 ng DNA per plasmid (Effectene, Qiagen, Hilden, Germany) into 96-well plates, grown for 24 h and assayed for Renilla and firefly luciferase activities. Experiments were performed in triplicate. All transcriptional activities were normalized relative to their intracellular concentration as determined by western blot analysis. The intracellular concentration itself was corrected for transfection efficiency on the basis of Renilla data. Samples for western blot analysis were transferred to a PVDF membrane (Immobilon-P 0.45 μM) (Millipore) using an XCell II blot module (Invitrogen, Karlsruhe, Germany). The blot was blocked in 5% skim milk and probed with mouse anti-myc antibody clone 4A6 (Millipore) or anti-GAPDH (Millipore). Detection was performed using an HRP goat anti-mouse IgG peroxide conjugate (Sigma, Hamburg, Germany). Blots were quantitated using the Biometra (Göttingen, Germany) BioDocAnalyze 2.0 software. Each experiment was repeated thrice.
Sequence alignments were performed with ClustalW2 (EBI, Cambridge, UK) and illustrated with Jalview (University of Dundee, UK).
Immunohistochemistry
Paraffin-embedded sections were dewaxed, rehydrated and stained with the 4A4 anti-p63 monoclonal or the 3A3 anti-p73 monoclonal antibody using standard protocols (MOM Immunohistochemistry Kit, Vector Laboratories, Peterborough, UK).
Accession numbers
The structure of p73 TD has been deposited at the Protein Data Bank (PDB) with the accession code 2KBY.
Accession codes
Abbreviations
- TD:
-
tetramerization domain
- OD:
-
oligomerization domain
- SAM:
-
sterile alpha motif
- CEP-1:
-
Caenorhabditis elegans p53
- Dmp53:
-
Drosophila melanogaster p53
- NMR:
-
nuclear magnetic resonance
- AUC:
-
analytical ultracentrifugation
- SE:
-
sedimentation equilibrium
- SV:
-
sedimentation velocity
- TROSY:
-
transverse relaxation-optimized spectroscopy
- NOE:
-
nuclear Overhauser effect
- NOESY:
-
nuclear Overhauser effect spectroscopy
- Gdn-HCl:
-
guanidine hydrochloride
- HNSCC:
-
head and neck squamous cell carcinoma
- TEV:
-
tobacco etch virus protease
- RDC:
-
residual dipolar coupling
References
Yang A, Kaghad M, Caput D, McKeon F . On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet 2002; 18: 90–95.
Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999; 398: 714–718.
Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A . p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999; 398: 708–713.
Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z et al. p63 protects the female germ line during meiotic arrest. Nature 2006; 444: 624–628.
Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 2000; 404: 99–103.
Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin Jr WG, Levrero M et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 1999; 399: 806–809.
Hu H, Xia SH, Li AD, Xu X, Cai Y, Han YL et al. Elevated expression of p63 protein in human esophageal squamous cell carcinomas. Int J Cancer 2002; 102: 580–583.
Massion PP, Taflan PM, Rahman SM, Yildiz P, Shyr Y, Carbone DP et al. Role of p63 amplification and overexpression in lung cancer development. Chest 2004; 125: 102S.
Sniezek JC, Matheny KE, Westfall MD, Pietenpol JA . Dominant negative p63 isoform expression in head and neck squamous cell carcinoma. Laryngoscope 2004; 114: 2063–2072.
Weber A, Bellmann U, Bootz F, Wittekind C, Tannapfel A . Expression of p53 and its homologues in primary and recurrent squamous cell carcinomas of the head and neck. Int J Cancer 2002; 99: 22–28.
Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW . p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell 2006; 9: 45–56.
Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998; 2: 305–316.
Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM . Drosophila p53 binds a damage response element at the reaper locus. Cell 2000; 101: 103–113.
Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S et al. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 2000; 101: 91–101.
Derry WB, Putzke AP, Rothman JH . Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science 2001; 294: 591–595.
Chene P . The role of tetramerization in p53 function. Oncogene 2001; 20: 2611–2617.
Shaulian E, Zauberman A, Ginsberg D, Oren M . Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol 1992; 12: 5581–5592.
Sturzbecher HW, Brain R, Addison C, Rudge K, Remm M, Grimaldi M et al. A C-terminal alpha-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene 1992; 7: 1513–1523.
Davison TS, Nie X, Ma W, Lin Y, Kay C, Benchimol S et al. Structure and functionality of a designed p53 dimer. J Mol Biol 2001; 307: 605–617.
Lomax ME, Barnes DM, Hupp TR, Picksley SM, Camplejohn RS . Characterization of p53 oligomerization domain mutations isolated from Li–Fraumeni and Li–Fraumeni like family members. Oncogene 1998; 17: 643–649.
Varley JM, McGown G, Thorncroft M, Cochrane S, Morrison P, Woll P et al. A previously undescribed mutation within the tetramerisation domain of TP53 in a family with Li–Fraumeni syndrome. Oncogene 1996; 12: 2437–2442.
Jeffrey PD, Gorina S, Pavletich NP . Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science 1995; 267: 1498–1502.
Lee W, Harvey TS, Yin Y, Yau P, Litchfield D, Arrowsmith CH . Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol 1994; 1: 877–890.
Jayaraman J, Prives C . Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell 1995; 81: 1021–1029.
Serber Z, Lai HC, Yang A, Ou HD, Sigal MS, Kelly AE et al. A C-terminal inhibitory domain controls the activity of p63 by an intramolecular mechanism. Mol Cell Biol 2002; 22: 8601–8611.
Ou HD, Löhr F, Vogel V, Mäntele W, Dötsch V . Structural evolution of C-terminal domains in the p53 family. EMBO J 2007; 26: 3463–3473.
De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M et al. Two new p73 splice variants, g and d, with different transcriptional activity. J Exp Med 1998; 188: 1763–1768.
Davison TS, Vagner C, Kaghad M, Ayed A, Caput D, Arrowsmith CH . p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J Biol Chem 1999; 274: 18709–18714.
Fernandez-Fernandez MR, Veprintsev DB, Fersht AR . Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc Natl Acad Sci USA 2005; 102: 4735–4740.
Rajagopalan S, Jaulent AM, Wells M, Veprintsev DB, Fersht AR . 14-3-3 activation of DNA binding of p53 by enhancing its association into tetramers. Nucleic Acids Res 2008; 36: 5983–5991.
Nyman U, Vlachos P, Cascante A, Hermanson O, Zhivotovsky B, Joseph B . Protein kinase C-dependent phosphorylation regulates the cell cycle-inhibitory function of the p73 carboxy terminus transactivation domain. Mol Cell Biol 2009; 29: 1814–1825.
Goddard TD, Kneller DG . SPARKY 3. University of California: San Francisco.
Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K . TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci USA 1998; 95: 13585–13590.
Salzmann M, Wider G, Pervushin K, Senn H, Wuthrich K . TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J Am Chem Soc 1999; 121: 844–848.
Lohr F, Hansel R, Rogov VV, Dotsch V . Improved pulse sequences for sequence specific assignment of aromatic proton resonances in proteins. J Biomol NMR 2007; 37: 205–224.
Cornilescu G, Delaglio F, Bax A . Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 1999; 13: 289–302.
Melacini G . Separation of intra- and intermolecular NOEs through simultaneous editing and J-compensated filtering: a 4D quadrature-free constant-time J-resolved approach. J Am Chem Soc 2000; 122: 9735–9738.
Bryce DL, Bax A . Application of correlated residual dipolar couplings to the determination of the molecular alignment tensor magnitude of oriented proteins and nucleic acids. J Biomol NMR 2004; 28: 273–287.
Ottiger M, Bax A . Determination of relative N-H-N N-C', C-alpha-C', and C(alpha)-H-alpha effective bond lengths in a protein by NMR in a dilute liquid crystalline phase. J Am Chem Soc 1998; 120: 12334–12341.
Linge JP, O'Donoghue SI, Nilges M . Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol 2001; 339: 71–90.
Acknowledgements
We thank Benjamin Bardiaux (Institut Pasteur) for providing a modified version of ARIA2.2 capable of handling tetramers with a D2 symmetry, and Hongmei Mou for help with immunohistochemistry. The research was funded by the Deutsche Forschungsgemeinschaft (DO 545/2-1), by the EU-Grant EPISTEM (LSHB-CT-019067), by the Centre for Biomolecular Magnetic Resonance at the University of Frankfurt (BMRZ), by the Cluster of Excellence Frankfurt (Macromolecular Complexes) and by the Volkswagen Foundation (PG). The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, from GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and from the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Edited by G Melino
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Coutandin, D., Löhr, F., Niesen, F. et al. Conformational stability and activity of p73 require a second helix in the tetramerization domain. Cell Death Differ 16, 1582–1589 (2009). https://doi.org/10.1038/cdd.2009.139
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2009.139
Keywords
This article is cited by
-
Disease-related p63 DBD mutations impair DNA binding by distinct mechanisms and varying degree
Cell Death & Disease (2023)
-
DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma
Cell Death & Disease (2023)
-
Structural diversity of p63 and p73 isoforms
Cell Death & Differentiation (2022)
-
Designed Ankyrin Repeat Proteins as a tool box for analyzing p63
Cell Death & Differentiation (2022)
-
p73 isoforms meet evolution of metastasis
Cancer and Metastasis Reviews (2022)