Abstract
WWP1 E3 ubiquitin ligase has previously been shown to be frequently amplified and overexpressed in prostate and breast cancers. However, the mechanism of WWP1 action is still largely unknown. p63, a member of the p53 family of transcription factors, has an important function in tumor development by regulating apoptosis. Using alternative promoters, p63 can be expressed as ΔNp63 and TAp63. Increasing evidence suggests that TAp63 sensitizes cells to apoptosis but ΔNp63 has an opposite function. In this study, we show that WWP1 binds, ubiquitinates, and destructs both ΔNp63α and TAp63α. The protein–protein interaction occurs between the PY motif of p63 and the WW domains of WWP1. The knockdown of WWP1 by siRNA increases the endogenous ΔNp63α level in the MCF10A and 184B5 immortalized breast epithelial cell lines and confers resistance to doxorubicin-induced apoptosis. On the other hand, the knockdown of WWP1 increases the endogenous level of TAp63α, induces apoptosis, and increases sensitivity to doxorubicin and cisplatin in the HCT116 colon cancer cell line in a p53-independent manner. Finally, we found that DNA damage chemotherapeutic drugs induce WWP1 mRNA and protein expression in a p53-dependent manner. These data suggest that WWP1 may have a context-dependent role in regulating cell survival through targeting different p63 proteins for degradation.
Similar content being viewed by others
Main
The WW domain containing E3 ubiquitin protein ligase 1 (WWP1) belongs to the C2-WW-HECT type E3 family, which comprises eight other members including NEDD4, AIP4/Itch, SMURF1, and SMURF2.1 All family members share a distinctive domain structure: a C2 domain at the N terminus for calcium-dependent phospholipid binding, 2–4 WW domains in the middle for protein–protein interaction with PY motifs, and a HECT domain at the C terminus for the ubiquitin transfer. Four family members including WWP1,2, 3 NEDD4,4 SMURF1,5 and SMURF2,6 have been shown to be overexpressed in different tumor types.
WWP1 is a potential oncogene that undergoes genomic amplification and overexpression in a subset of prostate and breast cancers.2, 3 The WWP1 gene is located at 8q21, a chromosomal region frequently amplified in human prostate and breast cancers. About 31–51% of cancer samples show gene copy number gains for WWP1. Furthermore, WWP1 is overexpressed in 58–60% of prostate and breast cancer samples. Functionally, WWP1 knockdown significantly suppresses cell proliferation and/or induces apoptosis in several prostate and breast cancer cell lines,2, 3 suggesting that WWP1 could be a promising molecular target for cancer therapy.
WWP1 is an intrinsic E3 ubiquitin ligase for multiple important proteins involved in tumorigenesis. The transforming growth factor-β signaling pathway is well known to suppress epithelial proliferation and induce apoptosis but promote tumor development at later stage. Several studies suggest that WWP1 negatively regulates the TGF-β signaling by targeting its molecular components, including TGF-β receptor 1 (TβR1),7 Smad2,8 and Smad49 for ubiquitin-mediated degradation. In addition, WWP1 has been reported to target several oncogenic factors such as Notch,10 Runx211 and KLF512 for ubiquitin-mediated proteolysis. Recently, WWP1 has been demonstrated to inhibit p53 activity through exporting p53 from the nucleus by ubiquitination.13 However, the role of WWP1 in tumorigenesis remains to be elucidated.
The p63 transcription factor, a member of the p53 family, shares DNA binding, oligomerization and possible transactivation (TA) domains with p53 and p73.14 Using alternative promoters, p63 can be expressed as ΔNp63 and TAp63 that have opposite functions in transcription control.15 There are three isoforms (α, β, γ) for both TAp63 and ΔNp63 because of the RNA splicing.16 All TAp63 isoforms contain an N-terminal p53-like transactivation domain which can transactivate traditional p53 target genes. Like p53, TAp63 promotes apoptosis through the death receptor and mitochondrial pathways. In contrast to TAp63, increasing evidence suggests that ΔNp63 isoforms could be oncoproteins with an anti-apoptotic activity. All ΔNp63 isoforms lack the TA domain but still contain the DNA-binding domain and oligomerization domain, suggesting that ΔNp63 may function as dominant-negative forms of TAp63. Indeed, the ΔNp63 isoforms have been shown to act as transcriptional repressors both in vitro and in vivo and strongly oppose the function of TAp63, TAp73 and p53.16 Numerous studies have shown that ΔNp63α is the predominate isoform expressed in epithelial tissues including epidermis, prostate and breast.16 p63 knockout mice fail to develop skin, prostate and mammary glands among other defects because of depletion of stem cells.17, 18 Similarly, inhibition of the endogenous ΔNp63α expression by RNAi induces epithelial apoptosis.19, 20 In addition, the ectopic expression of ΔNp63 in fibroblast cells induces anchorage-independent growth and tumor growth in nude mice.21
All p53 family members are degraded through the ubiquitin–proteasome pathway.22 Multiple E3 ligases including Mdm2, ARF-BP, Cop1 and Pirh2 have been demonstrated to target p53 for ubiquitin-mediated degradation.22 The receptor of activated protein C kinase-Elongin-C/B ubiquitin ligase complex has been proposed to be an E3 ligase for ΔNp63α.23 Recently, two WWP1 family members, Itch and Nedd4 have been shown to promote ubiquitination and degradation of the p63 proteins.24, 25 In addition, the inhibition of Itch potentiates the killing effect of doxorubicin in HeLa cells.26
Given the frequent expression alteration of WWP1 in human cancers, it is important to know whether WWP1 regulates apoptosis through promoting p63 degradation. Here, we show that WWP1 targets both ΔNp63α and TAp63α for ubiquitin-mediated proteasomal degradation. Importantly, we demonstrate that WWP1 regulates apoptosis and drug sensitivity in a p63-dependent manner. Lastly, we found WWP1 is induced by DNA damage therapeutic drugs in a p53-dependent manner. These findings help us understand the mechanism of WWP1 action in human cancer and may provide better designs for future cancer treatment.
Results
WWP1 interacts with p63α through the WW/PY motifs
The p63α protein has been shown to interact with Itch and Nedd4 through the PY/WW motif interaction.24, 25 To test whether WWP1 interacts with p63α, we first tested if p63α can be co-immunoprecipitated with WWP1. A plasmid expressing Myc-WWP1C886S (a catalytic inactive mouse WWP1 mutant) and plasmids expressing either FLAG-ΔNp63α or FLAG-TAp63α were transfected into LinX cells. Myc-WWP1C886S was efficiently immunoprecipitated by the anti-Myc antibody (Figure 1a). We found that both FLAG-ΔNp63α and FLAG-TAp63α are co-immunoprecipitated with Myc-WWP1C886S. The anti-Myc antibody itself cannot immunoprecipitate the p63α proteins without the expression of Myc-WWP1C886S, suggesting that the interactions are specific.
The WWP1 protein interacts with p63α proteins through the WW/PY motifs in mammalian cells. (a) Both FLAG-ΔNp63α and FLAG-TAp63α are co-precipitated with Myc-WWP1C886S. LinX cells were co-transfected with different combinations of expression plasmids for Myc-WWP1C886S, FLAG-ΔNp63α and FLAG-TAp63α. IP was performed using anti-Myc Ab. Myc-WWP1C886S was probed by anti-WWP1 Ab. The catalytic inactive WWP1 mutant was used to avoid p63 degradation by WT WWP1. β-actin serves as a loading control for the input. (b) Both FLAG-ΔNp63α and FLAG-TAp63α interact with WWP1 through the PY motif. FLAG-ΔNp63αY449F and FLAG-TAp63αY543F are two p63α mutants in which the PY motifs are disrupted by substituting the Tyr (Y) residue with the Phe (F) residue. The Myc-WWP1C886S-transfected LinX (without FLAG-p63α) was used as a negative control. (c) WWP1 binds to FLAG-ΔNp63α but not FLAG-ΔNp63αY449F through WW domains, as determined by GST pull-down assays. Four WW domains of WWP1 were individually or collectively expressed as GST fusion proteins in LinX cells. (d) The endogenous WWP1 protein forms a complex with the endogenous ΔNp63α protein in MCF10A. The WWP1 protein was probed with the rabbit anti-WWP1 antibody. The same amount of mouse IgG nonspecifically immunoprecipitated few ΔNp63α after extensive washing. However, anti-WWP1 Ab immunoprecipitated much more ΔNp63α than the IgG control under the same conditions. (e) The endogenous WWP1 protein forms a complex with the endogenous TAp63α protein in HCT116
Then we tested whether the protein–protein interaction between WWP1 and p63α is through the WW/PY motifs. We mutated the PY motifs in p63α (Y449F for ΔNp63α and Y543F for TAp63α) and performed immunoprecipitation (IP) with these mutants. We found that both PY motif-mutated p63α proteins cannot efficiently interact with Myc-WWP1C886S (Figure 1b), suggesting that the PY motifs in ΔNp63α and TAp63α are required to interact with WWP1. Following that, we asked which WW domain of WWP1 participates in the protein interaction with p63α. We fused each of the four WW domains of WWP1 to the C terminus of GST and performed GST pull-down assays with FLAG-ΔNp63α in LinX cells. As shown in Figure 1c, GST itself does not pull down any FLAG-ΔNp63α, but all four GST-WW proteins pull down different amounts of FLAG-ΔNp63α. The GST-WW1 protein pulls down much more FLAG-ΔNp63α than the rest of GST-WW proteins, although the expression levels of GST-WW and FLAG-ΔNp63α are similar in the different groups. Consistently, GST-WW (1–2) pulls down more FLAG-ΔNp63α than GST-WW (3–4). These findings suggest that the first WW domain may have a major function for p63α binding. Consistent with the results in Figure 1b, both GST-WW1 and GST-WW (1–2) pull down WT but not the PY motif-mutated ΔNp63α (Figure 1c, right panel). Taken together, the protein interaction between WWP1 and p63α is through the WW/PY motifs.
Finally, we immunoprecipitated the endogenous WWP1 proteins from MCF10A by using anti-WWP1 Ab and found that the endogenous ΔNp63α protein is in the same complex (Figure 1d). Similarly, we detected protein interaction between endogenous WWP1 and endogenous TAp63α in HCT116 (Figure 1e). These results suggest that the protein interaction between WWP1 and p63α could occur at the physiological level.
WWP1 ubiquitinates p63α in cultured mammalian cells
As the WWP1 E3 ligase interacts with both ΔNp63α and TAp63α, we next determined whether WWP1 ubiquitinates the p63α proteins in mammalian cells. To this end, we transfected the expression constructs for WT hWWP1 or the catalytic inactive mutant hWWP1C890A, FLAG-ΔNp63α or FLAG-TAp63α, and Myc-Ub into LinX cells. We performed IP with the anti-FLAG antibody conjugated M2 beads under a denaturing condition to eliminate any p63α-associated proteins through non-covalent bonds. The ubiquitin-conjugated p63α proteins were detected by western blot with anti-Myc Ab. As shown in Figure 2a, WT WWP1 significantly increases the ubiquitination of both TAp63α and ΔNp63α compared with the vector control, whereas the catalytic inactive WWP1C890A does not. We noticed that WWP1 shows a higher ubiquitination activity towards the ΔNp63 isoform compared with the TAp63 isoform. The format of p63 ubiquitination by WWP1 is most likely polyubiquitination because a smear of band above the unmodified p63α was detected for both TAp63α and ΔNp63α. These results indicate that WWP1 polyubiquitinates both ΔNp63α and TAp63α through its E3 ligase activity.
WWP1 ubiquitinates p63α proteins in mammalian cells. (a) WWP1 ubiquitinates both TAp63α and ΔNp63α using its E3 ligase activity. LinX cells were co-transfected with expressing plasmids for Myc-Ub, WWP1, WWP1C890A, FLAG-ΔNp63α and FLAG-TAp63α, as indicated. The cells were treated with 20 μM proteasome inhibitor MG132 overnight to accumulate the ubiquitinated p63α before harvest. The IP was performed with the anti-FLAG M2 beads under denaturing conditions. Immunoblotting was performed with the indicated Abs. The ubiquitin modified p63α proteins were detected by anti-Myc Ab. As majority of FLAG-ΔNp63α is polyubiquitinated by WWP1, much less unmodified FLAG-ΔNp63α was detected when WWP1 is co-transfected. (b) WWP1 specifically ubiquitinates WT but not PY motif-mutated p63α. (c) Endogenous WWP1 ubiquitinates endogenous ΔNp63α in MCF10A. WWP1 was knocked down by siRNA#1. Luc siRNA was used as a control. MG132 was not added. IP was performed under a denaturing condition using anti-p63 Ab and protein A beads
In addition, we examined the ubiquitination of the PY motif-mutated ΔNp63αY449F and TAp63αY543F by WWP1. As shown in Figure 2b, WWP1 only ubiquitinates WT p63α but not PY motif-mutated p63α under the same conditions. We conclude that the protein interaction is essential for WWP1 to ubiquitinate p63α.
To investigate whether endogenous WWP1 contributes to the endogenous ΔNp63α ubiquitination, we knocked down WWP1 in MCF10A by a WWP1 siRNA and examined the ubiquitination of ΔNp63α. Compared with the Luc siRNA, the WWP1 siRNA efficiently silenced the WWP1 protein expression, elevated the ΔNp63α protein level and decreased the ubiquitinated ΔNp63α.
WWP1 promotes p63α proteasomal degradation
To test whether WWP1 targets p63α for degradation, we first measured the steady-state levels of both TAp63α and ΔNp63α in the presence and absence of WWP1. As shown in Figure 3a, the steady-state levels of both TAp63α and ΔNp63α are decreased in WT WWP1, but not in WWP1C890A, overexpressing LinX cells. To further determine whether the protein interaction between p63α and WWP1 is required for degradation of p63α, we measured the steady-state protein level of ΔNp63αY449F in the presence of WWP1 or WWP1C890A. As expected, WT WWP1 fails to decrease the steady-state level of ΔNp63αY449F (Figure 3a).
WWP1 promotes proteasomal degradation of p63α proteins. (a) WT WWP1 decreases the steady levels of both FLAG-TAp63α and FLAG-ΔNp63α in LinX cells, as determined by western blot. The PY motif mutant FLAG-ΔNp63αY449F is resistant to WWP1-mediated degradation. An empty vector and the catalytic inactive mutant WWP1C890A were used as controls. (b) Measurement of protein half-lives by cycloheximide (CHX) chase assays and western blot. LinX cells were co-transfected with indicated plasmids. Forty-eight hours after transfection, the cells were incubated with 50 μg/ml CHX for different times (2–10 h) and collected for western blot. β-actin was used as a loading control. MG132 (20 μM) was added together with CHX as necessary. The exposure times have been adjusted for each panel to compare protein degradation. (c) Quantative results of ΔNp63α from panel B by the IMAGE J software. The normalized p63α at 0 h was defined as 100. V, vector; W, WWP1; Wm, WWP1C890A. (d) Quantative results of TAp63α from panel B by the IMAGE J software
To further investigate whether the decrease of p63α by WWP1 is because of the increase of protein degradation, we measured the half-lives of ΔNp63α and TAp63α in the presence and absence of WWP1 by cycloheximide (CHX) chase assays. As shown in Figure 3b, both the ΔNp63α and TAp63α proteins have a long half-life (>10 h) in LinX cells. When WT WWP1 is overexpressed, the half-lives are dramatically decreased to about 3.2 h for ΔNp63α (Figure 3c) and about 7.5 h for TAp63α (Figure 3d). The catalytic inactive WWP1C890A only slightly decreases the half-life of ΔNp63α and TAp63α when compared with the empty vector (Figure 3c and d). As the PY motif mutant p63α cannot interact with WWP1, we wondered whether WWP1 cannot decrease the half-lives of these mutant p63α. Indeed, we found that both ΔNp63αY449F and TAp63αY543F are resistant to WWP1-mediated degradation (Figure 3b–d).
The degradation of p63α by WWP1 is most likely through the proteasome as p63α is polyubiquitinated by WWP1. To test if the proteasome is involved in p63α degradation by WWP1, we performed CHX chase assays in the presence of the proteasome inhibitor MG132 (10 μM). As shown in Figure 3(b–d), WWP1-induced p63α degradation is blocked by MG132, suggesting that the degradation of p63α by WWP1 is through the 26S proteasome.
We also confirmed that endogenous WWP1 decreases protein half-life of endogenous ΔNp63α. The WWP1 protein was knocked down by siRNA in MCF10A, the ΔNp63α protein half-lives were measured by pulse chase assays. As shown in Supplementary Figure S1, WWP1 knockdown dramatically extended the half-life of the endogenous ΔNp63α protein.
WWP1 targets the endogenous ΔNp63α protein for degradation and sensitizes immortalized breast epithelial cells to chemotherapeutic drug doxorubicin-induced apoptosis
To determine whether WWP1 targets p63α under physiological conditions, we knocked down endogenous WWP1 in two immortalized breast epithelial cell lines MCF10A and 184B5 by two different anti-WWP1 siRNAs. We found that the protein levels of endogenous ΔNp63α, the major p63 isoform in MCF10A and 184B5, are remarkably elevated in both cell lines when WWP1 is knocked down by both siRNAs (Figure 4a). These results suggest that WWP1 targets the endogenous ΔNp63α protein for degradation.
WWP1 siRNA increases the endogenous ΔNp63α protein and confers resistance to doxorubicin in immortalized breast epithelial cells. (a) The knockdown of WWP1 by two different siRNAs increases the endogenous levels of the ΔNp63α protein in the MCF10A and 184B5 immortalized breast epithelial cells, as determined by western blot. siRNAs were transfected at 100 nM for 48 h. (b) WWP1 knockdown by siRNAs increases the cell viability in MCF10A cells. Different amounts of doxorubicin were added to cells 48 h after siRNA transfection. The cell viability was analyzed by the SRB assay 2 days later. Error bars mean S.D. (same for all figures in this paper). The data was collected from triplicate samples. The experiments were performed at least two times and similar results were obtained. (c) WWP1 knockdown by WWP1 siRNA#1 significantly decreases the doxorubicin (0.5 μM) induced apoptosis in MCF10A as determined by Annexin V staining. **P<0.01 (t-test). (d) WWP1 knockdown by WWP1 siRNA#1 decreases the doxorubicin-induced PARP and caspase 3 activation in MCF10A, as determined by Western blot. The cells were treated with doxorubicin for another 24 h after siRNA transfection for 48 h. The cleaved PARP is indicated by an arrow. (e) ΔNp63α siRNA rescues the WWP1 siRNA-induced doxorubicin resistance in MCF10A. All siRNAs were transfected at 50 nM for 72 h in total. All cells were treated with 0.25 μM doxorubicin for 48 h
It has been reported that ΔNp63α is essential for MCF10A to survive.19 Therefore, elevation of ΔNp63α may confer resistance to apoptosis. To test this, we treated siRNA-transfected MCF10A cells with different dosages of doxorubicin for 2 days and measured the cells viability by SRB assays. The high concentration of doxorubicin (⩾1 μM) kills all MCF10A cells at the same efficiency (Figure 4b). However, the WWP1 knockdown MCF10A cells are significantly more resistant to low concentrations (0.25–0.5 μM) of doxorubicin than the Ctrl-si-transfected cells. Two different anti-WWP1 siRNAs show similar results although Wsi#2 is slightly more effective than Wsi#1 possibly because of inducing more ΔNp63α (Figure 4a). We further measured apoptosis by the Annexin V staining. As shown in Figure 4c, Wsi#1 significantly (P<0.01) reduced doxorubicin (0.5 μM) induced Annexin V positive cells. We also confirmed apoptosis by the measurement of the cleaved PARP and caspase3 levels, two typical molecular markers for apoptosis. As shown in Figure 4d, the cleaved PARP and caspase3 are induced by doxorubicin (0.25–0.5 μM) in Ctrl-si-transfected MCF10A. In contrast, in Wsi#1-transfected MCF10A cells, the cleaved PARP and caspase3 are either undetectable or at low levels after treatment of doxorubicin (0.25–0.5 μM). Consistently, we noticed that the ΔNp63α protein levels are elevated in Wsi#1-transfected MCF10A cells without or with low concentration of doxorubicin. However, doxorubicin decreases the ΔNp63α protein levels in a dosage-dependent and WWP1-independent manner. The exact mechanism of ΔNp63α degradation by doxorubicin is currently unknown.
To further test whether WWP1 regulates apoptosis through targeting ΔNp63α for degradation, we knocked down both WWP1 and ΔNp63α in MCF10A and treated the cells with 0.25 μM doxorubicin (Figure 4e). Consistent with the previous results, the WWP1 siRNA-transfected MCF10A cells show a higher level of ΔNp63α and lower levels of cleaved PARP and cleaved caspase 3 than the Ctrl-si-transfected MCF10A cells. As reported,24 knockdown of ΔNp63α alone increases the levels of cleaved PARP and cleaved caspase 3. More importantly, knockdown of ΔNp63α abolishes the WWP1 siRNA-induced ΔNp63α increase and drug resistance. We also confirmed these results by the SRB assay (data not shown). These findings suggest that WWP1 regulates apoptosis and drug resistance in MCF10A through targeting ΔNp63α for degradation.
WWP1 targets the endogenous TAp63α protein for degradation and confers cell survival in a p53-independent manner
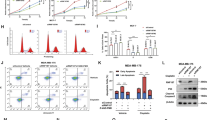
Based on the results in Figure 1, 2 and 3, WWP1 targets not only ΔNp63α but also TAp63α for degradation. Numerous studies have shown that the endogenous TAp63α protein is usually at low levels in epithelial cells.16 In a recent study, the endogenous TAp63α isoform is expressed in the HCT116 colon cancer cell line.27 We validated the expression of TAp63α in Figure 1e. Therefore, we sought to test whether WWP1 targets endogenous TAp63α in HCT116 cells and regulates apoptosis. As shown in Figure 5a, we efficiently knocked down WWP1 in HCT116 using two different anti-WWP1 siRNAs. As expected, the endogenous TAp63α levels are elevated in WWP1 knockdown cells. WWP1 siRNA#2 causes a more significant increase of TAp63α than siRNA#1 possibly because of different knockdown efficiency in this cell line. Then we measured apoptosis by PARP cleavage. Consistent with the elevated TAp63α levels, the cleaved PARP protein level is increased in the WWP1 knockdown HCT116 cells. We confirmed this result by measuring cell viability with SRB assays. We found that both anti-WWP1 siRNAs significantly decrease HCT116 cell viability (Figure 5b). Consistent with the higher levels of TAp63α and cleaved PARP, siRNA#2 more effectively decreases cell survival than siRNA#1. WWP1 has been shown to suppress p53 function.13 To further determine whether WWP1 regulates apoptosis through p53 rather than TAp63α, we performed the same experiment in p53-null HCT116 cells. There is no significant difference between the p53-null HCT116 cells and the p53 WT HCT116 cells in terms of the induction of TAp63α, cleaved PARP and apoptosis by WWP1 siRNA. We noticed that WWP1 siRNA may decrease cell viability slightly more effective in p53 WT HCT116 cells than in p53-null cells. However, the p53 levels are not significantly changed by WWP1 knockdown (Figure 5a). Furthermore, we measured apoptosis by Annexin V staining and found that WWP1 siRNA significantly increases Annexin V positive HCT116 cells (Figure 5c). These findings indicate that WWP1 siRNAs induce TAp63α and apoptosis in a p53-independent manner.
WWP1 knockdown by siRNAs upregulates the endogenous TAp63α levels and induces apoptosis in a p53-independent manner in HCT116 cells. (a) The knockdown of WWP1 by two different siRNAs increases the levels of the endogenous TAp63α protein and the cleaved PARP protein in the HCT116 and HCT116 (p53−/−) colon cancer cell lines. siRNAs were transfected at 100 nM for 48 h. (b) The knockdown of WWP1 by two different siRNAs decreases cell viability in the HCT116 and HCT116 (p53−/−) colon cancer cell lines. The cell viability was analyzed by the SRB assay after transfection with WWP1 siRNA for 72 h. *P<0.05; **P<0.01 (t-test). (c) WWP1 knockdown by WWP1 siRNA#2 significantly induces apoptosis in HCT116, as determined by Annexin V staining. *P<0.05 (t-test). (d) The knockdown of WWP1 in HCT116 cells decreases the IC50 for doxorubicin and cisplatin. The cell viability was analyzed by the SRB assay after transfection with WWP1 siRNA for 72 h in total and treatment with drugs for 48 h. The data was collected from triplicate samples. The experiments were performed at least two times and similar results were obtained
To study the biological relevance of WWP1-mediated degradation of TAp63α, we transfected WT TAp63α and the stable PY motif-mutated TAp63αY543F into HCT116 (p53−/−) cells and examined the proliferation/apoptosis index by SRB assays. We found that overexpression of TAp63α enhances cleaved caspase 3 and reduces cell viability when compared with the empty vector control (Supplementary Figure S2). Consistent with the protein stability, the level of TAp63αY543F is higher than that of WT TAp63α. Compared with WT TAp63, TAp63αY543F induces more caspase 3 cleavages and cell viability loss.
As WWP1 siRNA induces apoptosis in both HCT116 WT and p53-null cells, we further tested whether inhibition of WWP1 sensitizes cells to chemotherapeutic drugs. The HCT116 cells were transfected with Ctrl-siRNA or WWP1 siRNA for 1 day and treated with different concentration of doxorubicin or cisplatin for 2 days. As shown in Figure 5c, WWP1 siRNA and both chemotherapeutic drugs additively decrease the cell viability. After combination with WWP1 siRNA, the half maximal inhibitory concentration (IC50) for doxorubicin is decreased from 0.475 to 0.25 μM and the IC50 for cisplatin is decreased from 75 to 32.5 μM. Similar results were obtained in HCT116 p53-null cells (data not shown).
DNA damage chemotherapeutic drugs induces WWP1 expression in a p53-dependent manner
We noticed that the WWP1 protein is induced by doxorubicin in a dosage-dependent manner in MCF10A (Figure 4d). We wondered whether WWP1 is also induced by doxorubicin in HCT116 cells. To test this, the WT and p53-null HCT116 cells were treated with 1 μM doxorubicin at different times. As a result, doxorubicin induces both p53 and WWP1 in WT HCT116 cells in a time-dependent manner (Figure 6a). The induction peak for WWP1 is 48 h. Surprisingly, doxorubicin does not induce WWP1 in p53-null HCT116 cells. These results were conformed at mRNA levels by qRT-PCR (Figure 6b). These results indicate that doxorubicin may induce WWP1 transcription in a p53-dependent manner.
DNA damage drugs induce the WWP1 mRNA and protein in a p53-dependent manner. (a) The WWP1 protein is induced by doxorubicin (Dox, 1 μM) in a time-dependent manner in WT but not p53-null HCT116 cells. The WWP1, p53 and β-actin proteins were detected by western blot. (b) The WWP1 mRNA is induced by doxorubicin (Dox, 1 μM) in a time-dependent manner in WT but not p53-null HCT116 cells. The WWP1 and GAPDH mRNA were detected with qRT-PCR.3 (c) The WWP1 protein is induced by doxorubicin in a dose-dependent manner in WT but not p53-null HCT116 cells. (d) The WWP1 protein is induced by cisplatin (Cpt) in a dose-dependent manner in WT but not p53-null HCT116 cells. (e) The WWP1 protein is induced by doxorubicin in a dose-dependent manner in MCF10A (WT p53) but not in MDA-MB-468 (mutant p53) cells
To further test whether other DNA damage chemotherapeutic drugs also induce WWP1 in a p53-dependent manner, we treated the WT and p53-null HCT116 cells with different dosage of doxorubicin, cisplatin and etoposide. As shown in Figure 6c and d, both doxorubicin and cisplatin induce WWP1 in WT but not p53-null HCT116 cells in a dose-dependent manner. Similar results were observed with etoposide treatment (data not shown). These findings suggest that DNA damage chemotherapeutic drugs can induce the WWP1 expression in a p53-dependent manner. To further confirm these results in other cell lines other than HCT116, we treated MCF10A (WT p53)28 and MDA-MB-468 (mutant p53)29 with different concentration of doxorubicin for 48 h and found that WWP1 is only induced in MCF10A but not in MDA-MB-468 although p53 is accumulated in both cell lines (Figure 6e).
The protein expression of WWP1 and p63 is reversely correlated in prostate and breast cancer cell lines
The p63 protein has been documented to be specifically expressed in normal basal epithelial cells but lost in cancer cells from the prostate and breast.30, 31 In sharp contrast, WWP1 is frequently overexpressed in prostate and breast cancer cells. As WWP1 targets p63 for degradation, we asked whether the protein expression of WWP1 is negatively correlated with the protein expression of p63 in prostate and breast cancers.
We examined the protein expression levels of WWP1 and p63 in a panel of prostate and breast cell lines by western blot (Figure 7a–b). Consistent with our previous report,2 the protein levels of WWP1 are elevated in four prostate cancer cell lines LAPC-4, DU145, PC-3 and LNCaP/C4-2 compared with two immortalized prostate cell lines PZ-HPV7 and RWPE1 (Figure 7a). The protein expression of p63 (mainly ΔNp63α isoform) is negatively correlated with the expression of WWP1 in all these cell lines. The only exception is 22Rv1, in which the expression of WWP1 is not increased but p63 is still undetectable. Similar results are obtained in 10 breast cancer cell lines and four immortalized breast cell lines (Figure 7b). The only exception is HCC1937, in which the expression of WWP1 is increased but the ΔNp63 is still highly expressed. Therefore, we conclude that there is a negative correlation between WWP1 and p63 in a majority of prostate and breast cancer cell lines.
Expression of the WWP1 and p63 proteins in prostate and breast cell lines. Western blots were performed in a panel of prostate (a) and breast (b) cell lines to determine the protein expression levels of WWP1 and p63. The controls are immortalized cell lines. The molecular weight markers are labeled at the right. Tubulin was used as a loading control. The protein expression level of WWP1 is negatively correlated with the protein expression level of p63α, especially ΔNp63α (*) in these samples
Discussion
In this study, we provide several lines of evidence to support that WWP1 targets the p63 protein for ubiquitin-mediated proteasomal degradation. First, WWP1 binds to p63α proteins through the WW/PY motif interaction. Second, WWP1 ubiquitinates p63α through its E3 ligase activity. Third, WWP1 promotes p63α for proteasomal degradation. In addition, the WWP1 siRNA upregulates the endogenous ΔNp63α and TAp63α protein levels. Finally, there is a negative correlation between the WWP1 and p63 proteins in prostate and breast cells.
It is well documented that p63 proteins have important functions in epithelial development and tumorigenesis through regulating epithelial progenitor cell proliferation, differentiation and apoptosis.17, 18 p63 proteins are expressed in the basal cells of normal prostate glands but not in prostatic carcinomas.30 Similarly, the expression of p63 proteins is in the nuclei of myoepithelial cells of normal breast ducts and lobules but not in invasive breast cancer except metaplastic carcinomas.32 Several mechanisms including transcription regulation20 and post-translational modification contribute to the loss of p63 protein expression in prostate and breast cancers. Increasing evidence suggests that protein ubiquitination and degradation have an important function for the p63 activity.23, 24, 25, 33 Two other WWP1 family members, Itch and Nedd4, have been previously reported to target p63 proteins for proteasomal degradation.24, 25 However, the expression levels of Itch in breast cancer are not altered34 although the expression of Nedd4 has been found to be overexpressed in invasive bladder cancer cells.4 WWP1 is amplified and overexpressed in more than 30% of prostate and breast cancers, suggesting that WWP1 among these E3 ligases may have a major function for the p63 protein degradation in prostate and breast cancers, although we cannot completely exclude the roles of Itch and Nedd4. Under the physiological conditions, how WWP1, Itch and Nedd4 are coordinately activated and specially recruit p63 need further investigation.
Importantly, different p63 isoforms have different biological functions. There are at least six isoforms of p63, α, β and γ (each including TAp63 and ΔNp63). Numerous studies suggest that TAp63 isoforms induce apoptosis but ΔNp63 isoforms inhibit apoptosis. However, the functions of ΔNp63 from different reports are not consistent and sometimes even contradictory. ΔNp63 is essential for cancer cell survival through inhibiting TAp73 in squamous cell carcinoma (SCC)35 and a subset of breast cancer with p53 mutation.20 ΔNp63 has been reported to be overexpressed in primary SCCs of the head and neck.36 Thus, it would be interesting to examine whether WWP1 is frequently downregulated in SCCs. However, ΔNp63α is proposed to act as a metastasis suppressor by maintaining the epithelial phenotype of cancer cells.37 Loss of ΔNp63 has been reported to decrease epithelial cell adhesion and promote cell migration.19 Furthermore, ΔNp63 has even been demonstrated to possess a growth suppression function.38 p63 is frequently lost in most invasive adenocarcinomas including prostate cancer,30 breast cancer,31 bladder cancer39 and lung cancer.40 Therefore, the function of p63 may be context-dependent in different cancers.
It is well established that the functions of p63 depend on the expression pattern of p53 family members. In breast carcinomas, ΔNp63 isoforms are co-expressed with TAp73 exclusively within a subset of triple-negative (ER/EGFR/HER2 negative) primary breast cancers that commonly exhibit mutational inactivation of p53.20 We demonstrated that WWP1 regulates both ΔNp63α and TAp63α isoforms in this study. WWP1 may target the β isoforms but not the γ isoforms because both α and β, but not γ, have a PY motif. However, WWP1 has been reported to suppress the function of p53 through nuclear export although p53 does not have a PY motif.13 We found that WWP1 does not affect p53 stability in HCT116 and that WWP1 promotes p63 degradation independent of the p53 status (Figure 5a). In addition, Itch has been reported to target p73 for ubiquitin-mediated degradation because p73 contains PY motifs.41 Similarly, we found that WWP1 can also ubiquitinate p73 and decrease the stable level of p73 (unpublished observation). Thus, the functional output of WWP1 may rely on the expression pattern of p53 family members.
The expression pattern of p63 isoforms and p53 family members are different and complicated in different cancer cells. We found that WWP1 targets both ΔNp63α and TAp63α for degradation, implicating that WWP1 has a context-dependent role in terms of apoptosis and tumor development. Indeed, WWP1 sensitizes the MCF10A immortalized breast epithelial cells to doxorubicin-induced apoptosis but increases the HCT116 colon cancer cell survival and drug resistance. WWP1 is frequently amplified and overexpressed in prostate and breast cancers, suggesting that WWP1 overexpression may increase cell survival. In agreement with this idea, WWP1 knockdown in MCF7 and HCC1500 breast cancer cell lines causes apoptosis.3 We do not know whether the WWP1 ablation-induced apoptosis in these cells is actually through the increase of TAp63 because we did not detect the increase of TAp63 by western blot (data not shown). However, we cannot exclude that TAp63 has a function because TAp63 isoforms can be transcriptionally active at the levels below the limit of detection by western blot.42 Similarly, we cannot exclude that WWP1 promotes HCT116 cell survival through other molecules other than TAp63. The role and mechanistic action of WWP1 in cancer still need to be elucidated in vivo using transgenic mouse models.
WWP1 is amplified and overexpressed in over 40% of breast cancers and confers an advantage for cancer cell survival. In contrast, ΔNp63 is frequently lost in breast cancer cells. Apparently, the survival of the breast cancer cells with WWP1 overexpression does not depend on ΔNp63α. Only a small percentage of triple-negative breast cancers depend on ΔNp63 because TAp73 is overexpressed and p53 is mutated.20 If WWP1 inhibits TAp73 in these cells, the cell survival would not require ΔNp63 any longer. Therefore, we would predict that WWP1 expression is very low in triple-negative breast cancer cells. Indeed, WWP1 is preferably overexpressed in ER positive and p53 WT breast cancers (www.Oncomine.org). The immunohistochemical staining results in invasive breast tumors confirmed that the expression of WWP1 associates with positive ER status (Chen et al., manuscript submitted). Therefore, the expression of WWP1 may be invaluable for breast cancer diagnosis and prognosis.
The transcriptional regulation of WWP1 is largely unknown although we have previously shown that WWP1 is induced by TGFβ as a negative feedback mechanism.1 Laine et al.,13 reported that UV-irradiation or γ-irradiation decreases WWP1 mRNA expression in mouse embryonic fibroblasts in a p53-dependent manner. However, we found that several DNA damage chemotherapeutic drugs including doxorubicin, cisplatin and etoposide induces the WWP1 expression in HCT116 and breast cell lines in a p53-dependent manner. These conflicting results may be caused by different treatments in different cells. Whether WWP1 is an Mdm2 like p53 direct target gene needs elucidation in future studies.
In summary, we demonstrate that WWP1 targets p63 proteins for ubiquitin-mediated proteasomal degradation. We show that WWP1 plays opposite roles in terms of apoptosis and drug sensitivity in immortalized cells and cancer cells depending on the expression of different p63 isoforms. Given the frequent gene amplification and overexpression of WWP1 in prostate and breast cancers, these findings may help us understand the role of WWP1 in cancer development and may provide rationale to develop WWP1 as a diagnosis marker and molecular target for cancer therapy.
Materials and Methods
Cell culture and transfection
All breast and prostate cell lines have been described in our previous studies.3, 43, 44 The human embryonic kidney 293T-derived LinX cell line was cultured in DMEM containing 5% fetal bovine serum (FBS) and 1% penicillin and streptomycin (PS). The p53 wild type (WT) and p53-null HCT116 colon cancer cell lines were grown in HyQ®McCOY’S 5A medium with 5% FBS and 1% PS. All transient transfection for plasmids and siRNAs was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instruction. All chemically synthesized siRNAs were purchased from Dharmacon (Chicago, IL, USA) and transfected at 100 nM final concentration. The siRNA target sequences for human WWP1 gene are 5′-GAAGTCATCTGTAACTAAA-3′ (Wsi#1) and 5′-GCAGAGAAATACTGTTTAT-3′ (Wsi#2). The target sequence for ΔNp63α is 5′-CAGGTTGGCACTGAATTCACG-3′. The 3′-UU overhang for both strands and 5′-phophorylation for anti-sense sequence were employed for these siRNAs.
Expression plasmids
The plasmids expressing WT WWP1, the catalytic inactive hWWP1C890A, and mWWP1C886S have been described in our previous studies.3, 12 The Myc-Ub construct pCMV-6XHis-Myc-Ub is a gift from Dr. Raymond J. Deshaies (California Institute of Technology). The pcDNA3-FLAG-TAp63α and pcDNA3-FLAG-ΔNp63α constructs were obtained by subcloning the TAp63α and ΔNp63α cDNA into the Hind III and Not I sites of pcDNA3. The PY motif-mutated FLAG-TAp63αY543F and FLAG-ΔNp63αY449F were obtained by using a QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). Constructs for GST-WW domains were obtained by subcloning the DNA fragments encoding the WW domains of hWWP1 into the BamH I and Not I sites of the pEBG vector.
Immunoprecipitation and GST pull down
Immunoprecipitation using an anti-Myc antibody (Ab) plus protein A-agarose beads and GST pull down using the glutathione-Sepharose 4B slurry beads have been described in a previous study.12 Briefly, LinX cells were transfected with expression plasmids in 60-mm culture dishes for 48 h. The cells from each dish were collected into 0.6 ml of 1 × ice-cold cell lysis buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% protease inhibitor cocktail (no. P8340, Sigma, St. Louis, MO, USA) ) and incubated on ice for 30 min. Then, cell lysates were centrifuged at 10 000 g for 10 min at 4°C. The supernatant (200 μl) and primary anti-Myc antibody (2 μl) were incubated with gentle rocking overnight at 4°C. In all 30 μl of 50% protein A-agarose beads were added and incubated for 1–3 h at 4°C. The anti-WWP1 mouse monoclonal Ab (1A7) was used to replace anti-Myc Ab to immunoprecipitate the endogenous WWP1 proteins from MCF10A. The mouse IgG was used as the negative control. For the GST pull-down assay, the glutathione-Sepharose 4B slurry beads were directly incubated with the supernatant overnight at 4°C. The beads were washed five times with 500 μl of 1 × cell lysis buffer. Proteins were resuspended with 20–50 μl of SDS sample buffer and analyzed by western blot.
Antibodies and western blot
The anti-WWP1 rabbit polyclonal antibody (Ab) has been described in our previous report (3). The anti-WWP1 mouse monoclonal Ab (1A7) is from Novus Biologicals Inc. (Littleton, CO, USA). The anti-β-actin mouse monoclonal Ab AC-15 (no. A5441), the anti-FLAG rabbit polyclonal Ab (no. F7425), the anti-GST rabbit polyclonal Ab (no. G7781) are from Sigma. The anti-p63 mouse monoclonal Ab 4A4 (no. Sc-8431), anti-Myc mouse monoclonal Ab 9B11 (no. 2276), anti-PARP (no. 9915), anti-caspase 3 (no. 9915), and anti-tubulin antibodies (no. 2148) are from Cell Signaling (Danvers, MA, USA). The anti-p53 mouse monoclonal Ab (no. 554169) is from BD Pharmingen (San Diego, CA, USA).
Proteins were separated by SDS-PAGE and blotted onto PVDF membranes. The membranes were blocked with PBST (PBS with 0.1% Tween 20) buffer containing 5% nonfat dry milk and incubated with primary antibodies diluted in PBST overnight at 4°C. After washing three times in PBST, the membranes were incubated with 10 000 times diluted horseradish peroxidase-conjugated second antibodies for 1 h at room temperature. Detection was performed with the Supersignal West Pico enhanced chemiluminescence system (Pierce, Rockford, IL, USA) and a LAS-3000 Fujifilm imaging system.
Protein ubiquitination assay
LinX cells were transiently transfected with Myc-Ub and other plasmids as necessary in 6-well plates. Two days after transfection, the cells were harvested in 150 μl SDS lysis buffer (50 mM Tris-Cl, pH 6.8, 1.5% SDS). The samples were boiled for 15 min. 100 μl of protein lysate was diluted with 1.2 ml EBC/BSA buffer (50 mM Tris-Cl, pH 6.8, 180 mM NaCl, 0.5% CA630, 0.5% BSA) and incubated with 30 μl 50% anti-FLAG® M2-agarose beads (no. A2220, Sigma) overnight at 4°C with rotation. The beads were collected by centrifugation at 10 000 g for 30 s at 4°C and washed three times with 1 ml ice-cold EBC/BSA buffer. Proteins were resuspended with 30 μl of SDS sample buffer and analyzed by western blot. Ub-conjugated p63 was detected by anti-Myc Ab. For the endogenous p63 ubiquitination, IP was performed by using anti-p63 Ab; and ubiquitinated p63 was detected by anti-Ub Ab.
Measurement of apoptosis
The siRNA-transfected MCF10A and HCT116 cells were exposed to different concentrations of doxorubicin (Sigma) or Cisplatin (Cpt) at different times. The cell viability was measured by the SRB assay and the Annexin V staining as described in our previous report.2 The protein levels of cleaved PARP and/or cleaved caspase 3 were used to measure apoptosis.
Abbreviations
- Ab:
-
antibody
- CHX:
-
cycloheximide
- Cpt:
-
cisplatin
- DBD:
-
DNA-binding domain
- ER:
-
estrogen receptor
- HECT:
-
homologous to the E6-associated protein carboxyl terminus
- GST:
-
glutathione S-transferase
- IP:
-
immunoprecipitation
- RACK1:
-
receptor of activated protein C kinase
- SCC:
-
squamous cell carcinoma
- TA:
-
transactivation
- WT:
-
wild type
- WWP1:
-
WW domain-containing E3 ubiquitin protein ligase 1
References
Chen C, Matesic LE . The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev 2007; 26: 587–604.
Chen C, Sun X, Guo P, Dong XY, Sethi P, Zhou W et al. Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene 2007; 26: 2386–2394.
Chen C, Zhou Z, Ross JS, Zhou W, Dong JT . The amplified WWP1 gene is a potential molecular target in breast cancer. Int J Cancer 2007; 121: 2834–2841.
Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 2007; 128: 129–139.
Suzuki A, Shibata T, Shimada Y, Murakami Y, Horii A, Shiratori K et al. Identification of SMURF1 as a possible target for 7q21.3–22.1 amplification detected in a pancreatic cancer cell line by in-house array-based comparative genomic hybridization. Cancer Sci 2008; 99: 986–994.
Fukuchi M, Fukai Y, Masuda N, Miyazaki T, Nakajima M, Sohda M et al. High-level expression of the Smad ubiquitin ligase Smurf2 correlates with poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Res 2002; 62: 7162–7165.
Komuro A, Imamura T, Saitoh M, Yoshida Y, Yamori T, Miyazono K et al. Negative regulation of transforming growth factor-beta (TGF-beta) signaling by WW domain-containing protein 1 (WWP1). Oncogene 2004; 23: 6914–6923.
Seo SR, Lallemand F, Ferrand N, Pessah M, L’Hoste S, Camonis J et al. The novel E3 ubiquitin ligase Tiul1 associates with TGIF to target Smad2 for degradation. EMBO J 2004; 23: 3780–3792.
Moren A, Imamura T, Miyazono K, Heldin CH, Moustakas A . Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J Biol Chem 2005; 280: 22115–22123.
Shaye DD, Greenwald I . LIN-12/Notch trafficking and regulation of DSL ligand activity during vulval induction in Caenorhabditis elegans. Development 2005; 132: 5081–5092.
Shen R, Chen M, Wang YJ, Kaneki H, Xing L, O’Keefe R J et al. Smad6 interacts with Runx2 and mediates Smad ubiquitin regulatory factor 1-induced Runx2 degradation. J Biol Chem 2006; 281: 3569–3576.
Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X et al. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem 2005; 280: 41553–41561.
Laine A, Ronai Z . Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene 2007; 26: 1477–1483
Yang A, McKeon F . P63 and P73: P53 mimics, menaces and more. Nat Rev Mol Cell Biol 2000; 1: 199–207.
Wu G, Nomoto S, Hoque MO, Dracheva T, Osada M, Lee CC et al. DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res 2003; 63: 2351–2357.
Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V et al. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998; 2: 305–316.
Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999; 398: 714–718.
Senoo M, Pinto F, Crum CP, McKeon F . p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 2007; 129: 523–536.
Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol 2006; 8: 551–561.
Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW . The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest 2007; 117: 1370–1380.
Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci USA 2000; 97: 5462–5467.
Watson IR, Irwin MS . Ubiquitin and ubiquitin-like modifications of the p53 family. Neoplasia 2006; 8: 655–666.
Fomenkov A, Zangen R, Huang YP, Osada M, Guo Z, Fomenkov T et al. RACK1 and stratifin target DeltaNp63alpha for a proteasome degradation in head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle 2004; 3: 1285–1295.
Rossi M, Aqeilan RI, Neale M, Candi E, Salomoni P, Knight RA et al. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci USA 2006; 103: 12753–12758.
Bakkers J, Camacho-Carvajal M, Nowak M, Kramer C, Danger B, Hammerschmidt M . Destabilization of DeltaNp63alpha by Nedd4-mediated ubiquitination and Ubc9-mediated sumoylation, and its implications on dorsoventral patterning of the zebrafish embryo. Cell Cycle 2005; 4: 790–800.
Hansen TM, Rossi M, Roperch JP, Ansell K, Simpson K, Taylor D et al. Itch inhibition regulates chemosensitivity in vitro. Biochem Biophys Res Commun 2007; 361: 33–36.
Sayan BS, Sayan AE, Yang AL, Aqeilan RI, Candi E, Cohen GM et al. Cleavage of the transactivation-inhibitory domain of p63 by caspases enhances apoptosis. Proc Natl Acad Sci USA 2007; 104: 10871–10876.
Swaroop M, Sun Y . Mdm2 ligase dead mutants did not act in a dominant negative manner to re-activate p53, but promoted tumor cell growth. Anticancer Res 2003; 23: 3167–3174.
Liu W, Zhang R . Upregulation of p21WAF1/CIP1 in human breast cancer cell lines MCF-7 and MDA-MB-468 undergoing apoptosis induced by natural product anticancer drugs 10-hydroxycamptothecin and camptothecin through p53-dependent and independent pathways. Int J Oncol 1998; 12: 793–804.
Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L et al. p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol 2000; 157: 1769–1775.
Barbareschi M, Pecciarini L, Cangi MG, Macri E, Rizzo A, Viale G et al. p63, a p53 homologue, is a selective nuclear marker of myoepithelial cells of the human breast. Am J Surg Pathol 2001; 25: 1054–1060.
Koker MM, Kleer CG . p63 expression in breast cancer: a highly sensitive and specific marker of metaplastic carcinoma. Am J Surg Pathol 2004; 28: 1506–1512.
Rossi M, De Simone M, Pollice A, Santoro R, La Mantia G, Guerrini L et al. Itch/AIP4 associates with and promotes p63 protein degradation. Cell Cycle 2006; 5: 1816–1822.
Nguyen Huu N, Ryder W, Zeps N, Flasza M, Chiu M, Hanby A et al. Tumour-promoting activity of altered WWP1 expression in breast cancer and its utility as a prognostic indicator. J Pathol 2008; 216: 93–102.
Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW . p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell 2006; 9: 45–56.
Sniezek JC, Matheny KE, Westfall MD, Pietenpol JA . Dominant negative p63 isoform expression in head and neck squamous cell carcinoma. Laryngoscope 2004; 114: 2063–2072.
Barbieri CE, Pietenpol JA . p63 and epithelial biology. Exp Cell Res 2006; 312: 695–706.
Helton ES, Zhu J, Chen X . The unique NH2-terminally deleted (DeltaN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the DeltaN variant of p63. J Biol Chem 2006; 281: 2533–2542.
Urist MJ, Di Como CJ, Lu ML, Charytonowicz E, Verbel D, Crum CP et al. Loss of p63 expression is associated with tumor progression in bladder cancer. Am J Pathol 2002; 161: 1199–1206.
Massion PP, Taflan PM, Jamshedur Rahman SM, Yildiz P, Shyr Y, Edgerton ME et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res 2003; 63: 7113–7121.
Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH et al. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J 2005; 24: 836–848.
Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR . p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev 2004; 18: 126–131.
Chen C, Bhalala HV, Qiao H, Dong JT . A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene 2002; 21: 6567–6572.
Chen C, Bhalala HV, Vessella RL, Dong JT . KLF5 is frequently deleted and down-regulated but rarely mutated in prostate cancer. Prostate 2003; 55: 81–88.
Acknowledgements
This study was supported in part by a grant from American Cancer Society, A grant from The Department Of Defense Prostate Cancer Research Program (W81XWH-07-1-0191), and a Grant (BCTR0503705) from the Susan G Komen Breast Cancer Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by G Melino
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Rights and permissions
About this article
Cite this article
Li, Y., Zhou, Z. & Chen, C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ 15, 1941–1951 (2008). https://doi.org/10.1038/cdd.2008.134
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.134
Keywords
This article is cited by
-
Hypoxia-activated XBP1s recruits HDAC2-EZH2 to engage epigenetic suppression of ΔNp63α expression and promote breast cancer metastasis independent of HIF1α
Cell Death & Differentiation (2024)
-
The role of NEDD4 related HECT-type E3 ubiquitin ligases in defective autophagy in cancer cells: molecular mechanisms and therapeutic perspectives
Molecular Medicine (2023)
-
WWP1 E3 ligase at the crossroads of health and disease
Cell Death & Disease (2023)
-
The role of ubiquitin signaling pathway on liver regeneration in rats
Molecular and Cellular Biochemistry (2023)
-
Clinical significance of FBXW7 loss of function in human cancers
Molecular Cancer (2022)