Abstract
Background:
The objective of this study was to determine the molecular mechanisms responsible for cellular radiosensitivity in two human fibroblast cell lines 84BR and 175BR derived from two cancer patients.
Methods:
Clonogenic assays were performed following exposure to increasing doses of gamma radiation to confirm radiosensitivity. γ-H2AX foci assays were used to determine the efficiency of DNA double-strand break (DSB) repair in cells. Quantitative PCR (Q-PCR) established the expression levels of key DNA DSB repair genes. Imaging flow cytometry using annexin V-FITC was used to compare artemis expression and apoptosis in cells.
Results:
Clonogenic cellular hypersensitivity in the 84BR and 175BR cell lines was associated with a defect in DNA DSB repair measured by the γ-H2AX foci assay. The Q-PCR analysis and imaging flow cytometry revealed a two-fold overexpression of the artemis DNA repair gene, which was associated with an increased level of apoptosis in the cells before and after radiation exposure. Overexpression of normal artemis protein in a normal immortalised fibroblast cell line NB1-Tert resulted in increased radiosensitivity and apoptosis.
Conclusion:
We conclude that elevated expression of artemis is associated with higher levels of DNA DSB, radiosensitivity and elevated apoptosis in two radio-hypersensitive cell lines. These data reveal a potentially novel mechanism responsible for radiosensitivity and show that increased artemis expression in cells can result in either radiation resistance or enhanced sensitivity.
Similar content being viewed by others
Main
Efficient DNA repair mechanisms in human cells are central to the maintenance of genome integrity and the prevention of cancer. DNA double-strand breaks (DSB) are a potentially cytotoxic lesion and must be repaired by the cell to ensure survival and viability. DNA DSB can be generated by exposure to ionising radiation (IR) and certain classes of anticancer chemotherapeutic drugs, such as alkylating agents, but also as a consequence of normal cellular oxidative metabolism (Kryston et al, 2011). In addition, DSB can be caused by the action of recombinase activating gene-mediated VDJ recombination during the process of immunoglobulin production during adaptive immunity (Ramsden et al, 2010).
There are two principal cellular mechanisms which recognise and repair DSB. The canonical pathway of non-homologous end joining (NHEJ) functions primarily in non-cycling cells or in the early phases of the cell cycle (Rothkamm et al, 2003). During NHEJ, the DNA-dependent protein kinase heterodimer, which contains the Ku70 and Ku80 protein complex, binds to DNA ends and recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to the break site (West et al, 1998). Due to the fact that the DNA ends at the break are rarely compatible, DNA-PKcs recruits the artemis protein, which acquires endonucleolytic activity and digests 5′ and 3′ DNA overhangs. The compatible DNA ends are re-ligated by a protein complex containing DNA ligase IV, which is stabilised and stimulated by XRCC4 and XLF-cernunos (Burma et al, 2006).
Animal models reveal that defects in any component of the NHEJ pathway can lead to hypersensitivity to IR, genome instability, immunodeficiency and cancer (Bassing and Alt, 2004). In humans, defects in the specific genetic components of NHEJ also result in predisposition to cancer (lymphoma and leukaemia) or to extreme radiosensitivity. For example, defects in ligase IV (Riballo et al, 1999), deficiency in artemis expression (Musio et al, 2005) or a mutation in the DNA-PKcs gene are associated with extreme clinical and cellular radiosensitivity and increased cancer incidence (Abbaszadeh et al, 2010, van der Burg et al, 2010).
Alternatively, in cells traversing the later phases of the cell cycle, such as the S phase, DSB repair is mediated primarily by the homologous recombination (HR) pathway in an error-free manner (Scott and Pandita, 2006). A Holliday junction is created, whereby lost genetic information is ‘copied’ from the homologous sister chromatid. Following resolution and separation of the two sister chromatids by the action of resolvase enzymes, DNA replication is completed and genomic integrity at the site of the DSB is maintained (Ip et al, 2008).
Defects leading to a reduction in functional artemis endonuclease activity are particularly noteworthy. At the cellular level, this manifests as extreme radiosensitivity. In human subjects, artemis deficiency results in radiation-sensitive severe combined immunodeficiency A (SCID-A). Severe combined immunodeficiency A is characterised clinically by extreme radiosensitivity and by a complete absence of T and B cells due to a failure of the receptor recombination stage of V(D)J recombination (Dvorak and Cowan, 2010). Attempts to correct the deficiency in cells by transduction with lentiviral gene expression vectors containing the artemis cDNA has led to overexpression of artemis in a number of mammalian cell types (Multhaup et al, 2010). However, overexpression has also resulted in a loss of cell viability, associated with increased DNA damage and elevated apoptosis as a result of abnormal activity of the artemis endonuclease inducing DNA breaks (Multhaup et al, 2010). Although such findings have significant implications for the role of gene therapy approaches for the treatment of SCID, it also reveals a potentially novel mechanism, which may explain radiosensitivity in human cell types. To support this assertion, we have recently characterised two radiosensitive human fibroblast cell lines derived from different cancer patients in which we observed a two-fold increase in the expression of artemis as determined by real-time quantitative PCR (Q-PCR) analysis. This was also associated with enhanced cellular radiosensitivity, together with a reduction in the repair of DNA DSB and increased levels of apoptosis in both cell lines. Also, in an immortalised normal fibroblast cell line NB1-Tert transfected to overexpress the artemis protein, we observed increased cellular radiosensitivity in a clonogenic assay following exposure to a range of doses of gamma radiation. From these observations, we propose a novel mechanism of cellular radiation hypersensitivity, whereby abnormally high expression of artemis may act as a dominant negative resulting in increased DNA DSB and elevated apoptosis associated with clinical and cellular radiosensitivity.
Materials and methods
Cell culture
The 84BR fibroblast cell line was derived from a non-affected skin biopsy of radiosensitive breast cancer patient (Arlett et al, 1989). The 175BR cell line was derived from a non-affected skin biopsy from an individual with multiple tumours of independent histological origin and was a kind gift from Professor Bruce Ponder (Cambridge Research Institute, Cambridge, UK). The NB1 normal fibroblast cell line is described by Bridger and Kill (2004) and the 1.BR3 fibroblast cell line was derived from a normal individual (Arlett et al, 1988). The NB1-Tert cell line was derived from the NB1 cells described above but were immortalised by transfection with the human HTERT cDNA using the standard protocols described elsewhere (Hahn et al, 1999). Cells were routinely cultured in Dulbecco’s Modified Eagle Medium (DMEM) (PAA Laboratories Ltd, Yeovil, UK), supplemented with 10% (v/v) fetal calf serum (PAA Laboratories Ltd), 2 mM L-glutamine and 100 Uml−1 penicillin and streptomycin (PAA Laboratories Ltd). Cells were grown as monolayers at 37 °C in a humidified atmosphere of 5% CO2 in air.
Clonogenic cell survival assays
Radiation
Clonogenic assays following exposure to gamma radiation were conducted as described previously (Arlett et al, 2006). In brief, cells were irradiated in suspension in DMEM with 0, 2, 4, 6 and 8 Gy gamma radiation from a 60Cobalt source at a distance of 25 cm from the source at a dose rate of 1.4–1.5 Gy min−1. An appropriate number of cells were plated into each of five 10-cm dishes, containing 10 ml DMEM as described above, to allow the growth of 200–300 colonies in untreated control dishes. Cells were incubated for a period of 3 weeks, fixed in methanol, stained with 0.5% methylene blue in water (Sigma-Aldrich Ltd, Dorset, UK) and colonies counted. The number of colonies derived from irradiated cells was expressed as a percentage of colonies in untreated control plates.
Nitrogen mustard clonogenic assays
Clonogenic assays following exposure of cells to nitrogen mustard (HN2) was conducted as described by Clingen et al (2007). In brief, approximately 1 × 105 cells were seeded into 5-cm dishes in 5 ml complete medium and incubated overnight. Analytical grade HN2 (Sigma Aldrich Ltd) was diluted immediately before use in serum-free medium. Cells were exposed to HN2 for 1 h, after which the cells were washed in serum-free medium, trypsinised and counted and inoculated into five 10-cm dishes per dose, containing 10 ml complete medium. Colonies formed after a 3-week incubation period were fixed, stained and counted as described for the radiation exposure clonogenic assays.
Determination of DNA DSB using γ-H2AX
The repair of DNA DSB following exposure to 2 Gy gamma radiation from a 60Cobalt source was measured as described previously (Abbaszadeh et al, 2010). Cells were grown as monolayers on 13-mm diameter circular coverslips. A monoclonal antibody to phosphoserine139 γ-H2AX, clone JBW 301 (Millipore Ltd, Watford, UK), at 1 : 10 000 dilution was added for 1 h at room temperature (RT). Alexa Fluor 488 goat antimouse IgG antibody (Invitrogen Ltd, Paisley, Scotland, UK) diluted 1 : 1000 was added to the cells for 1 h at RT, after which cells were counterstained with 1 μg ml−1 4′,6-diamidino-2-phenylindole in mowiol mountant (Sigma Aldrich Ltd). Analysis was performed by counting the number of foci in a minimum of 200 nuclei of each cell line at each time point (unirradiated and 30 min, 3, 5 and 24 h post exposure) using a Zeiss Axioscope fluorescence microscope with a × 100 magnification objective (Carl Zeiss Ltd, Cambridge, UK).
Apoptosis assay
The level of apoptosis in untreated cells and cells irradiated with 2 Gy gamma radiation was determined by immunocytochemistry and imaging flow cytometry, using a FITC Annexin V Apoptosis detection kit (BD Pharmingen Ltd, Oxford, UK) according to the manufacturer’s instructions. In brief, either untreated cells or cells irradiated with 2 Gy gamma radiation were recovered into suspension by trypsinisation and washed twice in PBS. Cells were resuspended into 500 μl of binding buffer, to which was added 5 μl annexin V-FITC and 5 μl of propidium iodide (PI). Cells were incubated for 10 min at RT in the dark and levels of apoptosis were determining by counting 5000 cells using the ImagestreamX imaging flow cytometer with a 488-nm laser at a power setting of 40 mW (Amnis Corporation, Seattle, WA, USA). Images of all cells were visualised using the Ideas analysis software program of the ImagestreamX (e.g., Bourton et al, 2011). Here cells are first gated for single cells, cells in focus, followed by identification of apoptotic cells. Apoptotic cells were gated and enumerated by identifying those cells that exhibited FITC and PI staining. Staining patterns in all cells were visually confirmed by assessing the appearance of the cells using the Image Gallery of the Ideas software package.
Total RNA extraction and cDNA synthesis
Total RNA was isolated from a single 10-cm dish of 80% confluent 1BR.3, NB1, 175BR and 84BR cells using Trizol reagent (Sigma Aldrich Ltd) according to the manufacturer’s instructions. Total RNA was quantified by spectrophotometry and visually evaluated by agarose gel electrophoresis. Reverse transcription of the RNA into first-strand cDNA was achieved by using 1 μg of total RNA from each cell line using Superscript III reverse transcriptase (Invitrogen Ltd) and random hexamer primers (Invitrogen Ltd).
Real-time Q-PCR
A SYBR green fluorescence-based real-time Q-PCR assay was used for the determination of the expression levels of all key genes involved in NHEJ DNA DSB repair. Reactions were carried out using an ABI Prism 7900HT Fast Real-Time PCR system (Applied Biosystems Ltd, Paisley, Scotland, UK). Using the NCBI Sequence Database, gene-specific primers were designed and synthesised for Ku70, Ku80, DNA-PKcs, XRCC4, ligase IV, XLF-cernunnos and artemis. The sequences for both forward and reverse primers, together with amplicon sizes, are shown in Table 1.
A 20-μl reaction volume included 10 μl (2 × ) SYBR green mastermix (Applied Biosystems Ltd), 1 μl of forward and reverse primer (10 μ M concentration) and 50 nM cDNA. Reaction volumes were made up to 20 μl using molecular biology grade water.
NHEJ target genes and GAPDH or β-actin were amplified in triplicate reactions for each sample and the average CT values were calculated. Relative quantification analysis of gene expression was carried out using the comparative CT (2−ΔΔCT) method (Livak and Schmitten, 2001) for the determination of expression of all NHEJ genes. GAPDH or β-actin expression were used as the internal reference genes for the normalisation of NHEJ gene expression. The NB1 and 1BR.3 normal fibroblast cell lines were used as calibrators.
Transfection of NB1-Tert cells with the artemis cDNA
The artemis cDNA expression construct was purchased from Origene Inc. (Rockville, MD, USA), which was provided in the expression plasmid pCMV6-XL5 (under control of the CMV promoter). This plasmid was supplied without a selectable marker to identify transfected cells. Therefore, the NB1-Tert cells were co-transfected with the pCMV6-XL5 and pPur plasmid (Clontech, Hants, UK). pPur confers resistance to puromycin allowing the selection of transfected cells.
Transfection was conducted using Genejuice lipid transfection reagent (Novagen Ltd, London, UK). A total of 20μg of each plasmid in 100 μl of serum-free medium containing 3 μl of Genejuice reagent was added to the cells for 5 h, after which cells were selected in 20 μg ml−1 puromycin for 4 weeks. Puromycin-resistant clones were collected by ring cloning and subjected to further analysis.
Quantitation of artemis protein expression in cells
Artemis protein expression in the fibroblast cell lines NB1, 1BR.3, 84BR and 175BR and the NB1-Tert cells transfected with artemis was determined using immunocytochemistry and imaging flow cytometry. Cells growing as monolayers were trypsinised and washed twice in 10 ml of ice cold PBS. Following fixation in 3.7% paraformaldehyde in PBS at 4°C for 15 min, cells were washed in PBS and fixed in 50 : 50 vol:vol methanol acetone for 10 min at 4 °C. Cells were rehydrated and permeabilised in Tris-buffered saline containing 0.1% Tween 20, after which the cells were stained with a rabbit polyclonal anti-artemis antibody at a dilution of 1 : 100 in permeabilisation buffer containing 5% goat serum (Abcam, Cambridge, UK). Following two washes in permeabilisation buffer, a goat anti-rabbit Alexa Fluor-conjugated secondary antibody was added at a dilution of 1 : 1000 for 1 h at RT (Invitrogen Ltd). Cells were washed three times in permeabilisation buffer and resuspended in 100 μl Accumax flow cytometry buffer (PAA Laboratories Ltd) containing 5 μ M Draq 5 (to visualise the nuclear region). Images of 10 000 cells were captured using the ImagestreamX, and the level of artemis expression in cells was determined by calculating the average Alexa Fluor-associated fluorescence in the nuclei of the cells.
Statistical analysis
Student’s unpaired t-tests were used to compare the expression of key NHEJ genes in the radiation-sensitive 84BR and 175BR fibroblast cell lines and NB1 and 1BR.3 normal fibroblast cell lines. In addition, the difference in the survival of cells following both HN2 and gamma radiation exposure was also compared using Student’s unpaired t-tests.
Results
Clonogenic cell-survival assays – radiation
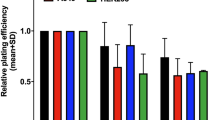
The response of the 84BR and 175BR fibroblast cell lines to increasing doses of gamma radiation is shown in Figure 1 and is compared with the response from the repair normal human fibroblast cell lines NB1 and 1BR.3. It can be seen for all cell lines that exposure to increasing doses of gamma radiation causes a proportionate decrease in clonogenic cell survival. However, in the 84BR and 175BR cell lines, there is greater reduction in colony survival when compared with the NB1 and 1.BR3 normal fibroblast cell lines. Comparison of the D10 values (the dose of radiation required to reduce survival to 10%) show that the D10 values for the repair normal fibroblasts is 5 Gy for the NB1 cells and 5.7 Gy for the 1BR.3 cells. Therefore, the NB1 cells are 1.47-fold more radioresistant than the 84BR and the 175BR cells, whereas the 1BR.3 cells are 1.68-fold more radioresistant than the 84BR and 175BR cell lines. These observations confirm those of Arlett et al 1989 and demonstrate the inherent cellular radiosensitivity of the 84BR and 175BR fibroblast cell lines. A Student’s unpaired t-test, comparing the D10 values of the 84BR and 175BR cell lines with the normal fibroblasts, demonstrated that the cells were significantly more sensitive to gamma radiation (P<0.05).
The clonogenic cell survival of the radiosensitive 84BR and 175BR cell lines in comparison with the repair normal fibroblasts 1BR.3 and NB1 following exposure to increasing doses of gamma radiation. Data are derived from at least three independent experiments, and error bars represent s.e. of the mean survival following exposure to 0, 2, 4, 6, and 8 Gy gamma radiation.
Clonogenic cell-survival assays – HN2
DNA DSB can be repaired by one of the two mutually exclusive repair pathways as described above. In order to determine if the radiosensitivity of the 84BR and 175BR cell lines was associated with a defect in either the NHEJ- or HR-repair pathway, clonogenic assays were performed where cells were exposed to increasing concentrations of the chemotherapeutic drug HN2. This agent primarily exerts its cytotoxicity by the introduction of DNA interstrand cross-links (ICL), which compromise both strands of the DNA. Such adducts are particularly challenging for the cell to repair, which occurs via a two-step process. Here the ICL is ‘unhooked’ from one strand via the action of the nucleotide excision repair-associated endonuclease ERCC1–XPF protein complex. This in turn creates a substrate for repair of the DSB by HR. Figure 2 shows the response of the radiosensitive 84BR and 175BR cell lines, together with the repair normal NB1 and 1BR.3 cells. It can be observed that for all cell lines as the concentration of HN2 increases, there is a decrease in clonogenic cell survival. However, all cell lines display a similar sensitivity. Using a Student’s unpaired t-test to compare the IC50 values of the 84BR and 175BR cell lines with the normal fibroblasts, it was demonstrated that there were no significant differences in IC50 values, indicating similar responses to HN2 (P=0.18). This observation indicates that the radiosensitivity of the 84BR and 175BR cell lines is not due to a defect in the HR-repair pathway and more likely to be a defect in one or more components of the NHEJ-repair pathway.
The clonogenic cell survival of the radiosensitive 84BR and 175BR cell lines in comparison with the repair normal fibroblasts 1BR.3 and NB1 following exposure to increasing doses of the cross-linking chemotherapeutic drug nitrogen mustard. Data are derived from a minimum of three independent experiments, and error bars represent s.e. of the mean survival at each concentration of drug.
γ -H2AX foci assay: DNA DSB repair
The induction and repair of DNA DSB in all cell lines was determined by counting the number of γ-H2AX foci in the nuclei of untreated cells and those exposed to 2 Gy gamma radiation at 30 min, 3, 5 and 24 h post irradiation. The data displayed in Figure 3 show that in the untreated cells, there are more residual γ-H2AX foci in the radiosensitive 84BR and 175BR cell lines (5.12 and 4.32 foci per nucleus, respectively), compared with the repair normal NB1 and 1BR.3 cells (0.81 and 1.02 foci per nucleus, respectively). At 30 min post irradiation, there is a dramatic induction of foci in all cells consistent with the induction of DNA DSB. In the NB1 and 1BR.3 cells, the number of foci return to near normal levels at 5 and 24 h. In the 84BR and 175BR cells, however, there is a significant retention of foci at 5 and 24 h when compared with the NB1 and 1BR.3 cells (P<0.05).
The induction and repair of DNA DSB in the radiosensitive 84BR and 175BR fibroblasts in comparison with the repair normal 1BR.3 and NB1 fibroblast cells. Cells were exposed to 2 Gy gamma radiation, and the induction and repair of DNA DSB was measured by the appearance and removal of γ-H2AX foci in the nuclei of unirradiated and irradiated cells. Data are derived from three independent experiments, and error bars represent s.e. of the mean.
Expression of NHEJ genes: Q-PCR
The levels of expression of the key genes involved in NHEJ for both radiosensitive 84BR and 175BR cells lines are shown in Figure 4A and B. In each experiment, the repair normal NB1 and 1BR.3 fibroblasts are presented as internal calibrators for the expression of the NHEJ genes, and the levels of expression in these cells are represented by a value of 1.0. Expression of the NHEJ genes in the radiosensitive cells is plotted relative to the repair normal fibroblasts. For the 84BR fibroblasts, there is an approximate two-fold overexpression of the artemis protein when compared with the repair normal NB1 and 1BR.3 fibroblasts. Comparison of the expression levels of artemis in the 84BR cells with the repair normal fibroblast using a Student’s unpaired t-test reveals a statistically significant difference in expression levels (P=0.003). Similarly for the radiosensitive 175BR fibroblasts, it can be seen that there is an approximate 1.7-fold overexpression of the artemis protein when compared with the control cells. A Student’s unpaired t-test reveals a statistically significant difference in expression levels between the 175BR radiosensitive cells and the repair normal control cells (P=0.007).
(A) The expression of NHEJ genes in the radiosensitive 84BR fibroblasts in comparison with the repair normal NB1 and 1BR.3 cells is shown. The data are derived from three independent experiments, and error bars show s.e. of the mean. Gene expression in the 84BR cells is expressed in comparison with the normal fibroblasts cells (calibrators), in which levels of expression are regarded as a level of 1. (B) The data for the 175BR radiosensitive cells are shown and presented in an identical manner to that of Figure 4A.
Radiation sensitivity of artemis-transfected NB1-Tert cells
Clonogenic cell survival in the artemis-transfected NB1-Tert cells following exposure to 0, 2, 4, 6 and 8 Gy gamma radiation is shown in Figure 5. In the parental NB1-Tert cells and those transfected with the pPur plasmid, together with a single clone (clone 6) of artemis-transfected cells, there is a similar and normal level of survival. However, in the NB1-Tert clones 1–5 (that are overexpressing artemis), there is a marked reduction in the clonogenic cell survival following exposure to increasing doses of gamma radiation. In the single clone 6 in which the radiation response was unaltered, it is likely that here the cells were transfected with the pPur plasmid only and failed to acquire the artemis gene during the co-transfection process.
The survival of the NB1-Tert cells transfected with and overexpressing the artemis endonuclease following exposure to 0, 2, 4, 6 and 8 Gy gamma radiation is shown. In clones 1–5, clonogenic cell survival is reduced by overexpression of the artemis gene. Cell survival in the vector-only transfected cells is unchanged and similar to the parental cells. A single clone (clone 6) of NB1-Tert-transfected cells also shows unchanged radiation sensitivity. This is likely because of the failure of the artemis transfection during the co-transfection procedure. Data are derived from three independent experiments, and error bars represent s.e. of the mean.
Analysis of apoptosis levels in the fibroblast cell lines and NB1-Tert artemis-transfected cell lines
Apoptosis in NB1, 1BR.3, 84BR and 175BR cells
Using annexin V and PI staining of live cells in conjunction with ImagestreamX imaging flow cytometry, the levels of apoptosis in unirradiated and cells exposed to 2 Gy gamma radiation was assessed in approximately 5000 cells for each cell line. The data shown in Figure 6A show that in the repair normal NB1 and 1BR.3 cells, the levels of apoptosis in unirradiated cells (determined by the number of cells positive for annexin V and PI staining) is 5.00% and 3.31% of all cells, respectively. Following 2 Gy gamma radiation, the levels of radiation-induced apoptosis increases to 8.65% and 11.50%, respectively, for each cell line. In the radiation-sensitive 84BR and 175BR fibroblasts, apoptosis levels in unirradiated cells is 8.65% and 10.32%, respectively. Following 2 Gy irradiation, these levels increase to 24.54% and 30.60%, respectively. These data indicate that spontaneous levels of apoptosis in the radiosensitive cell lines are higher than in the repair-competent NB1 and 1BR.3 cells. Moreover, a similar and significantly increased level of apoptosis is observed in the 84BR and 175BR cells compared with the repair-competent fibroblasts (P<0.05, Student’s unpaired t-test). In Figure 7A, representative images of cells in apoptosis are shown.
(A) Apoptosis levels in the NB1 and 1BR.3 repair normal cells compared with the radiosensitive 84BR and 175BR cells prior to (white bars) and following 2 Gy gamma radiation (grey bars) are shown. Apoptosis was determined by annexin V and PI staining, and data are derived from imaging flow cytometry using the Imagestream, whereby images of ⩾5000 cells were captured and apoptotic cells determined. The s.d. bars are included and shown on the graphs. (B) The levels of apoptosis in the NB1-Tert cells transfected to overexpress the artemis protein are shown. Data are derived from analysing the images of not less than 5000 cells. The s.d. bars are included and shown on the graphs. White bars are untreated cells. Grey bars are cells treated with 2 Gy gamma radiation.
(A) Representative examples of non-apoptotic and cells at different stages of apoptosis, derived from Imagestream analysis. A brightfield image is shown together with an image of annexin V staining (FITC) and an image of the nucleus stained with PI. Rows a–d show cells at advancing stages of apoptosis. (B) Representative images of NB1-Tert cells, which have been stained for artemis. A brightfield image is shown in the left column. The centre column shows Alexa Fluor staining for artemis and the right column shows Draq 5 staining of the nuclear region of the cell. Row a shows NB1-Tert parental cells. Row b shows elevated artemis expression in NB1-Tert clone 2, which has been transfected to overexpress the artemis protein.
Elevated apoptosis in artemis-transfected NB1-Tert cells
Apoptosis in NB1-Tert cells transfected to overexpress artemis was measured by imaging flow cytometry. In five out of six clones, we demonstrated a significantly increased level of apoptosis when compared with non-transfected cells and cells transfected with the pPur vector only (P<0.05). In clone 6, we did not observe elevated apoptosis. Interestingly, this clone was not observed to have elevated artemis expression (Table 2); this is likely because of the failure to transfect with the artemis cDNA during co-transfection.
Artemis expression in cells
The level of artemis expression in cells was measured by the method of imaging flow cytometry in conjunction with immunocytochemistry. Cells growing as monolayers were trypsinised and stained with antibodies to detect artemis. Images of 10 000 cells were captured with imaging flow cytometry, and using a series of pre-determined analysis routines in the Ideas software (Amnis Inc.) (e.g., Bourton et al, 2011), the average level of nuclear fluorescence attributable to artemis expression was calculated. The data are summarised in Table 2.
In the NB1 and 1BR.3 normal fibroblast cells, the average nuclear fluorescence attributable to artemis expression was averaged and given a value of 1.0. Expression of artemis in the 84BR and 175BR was expressed in relation to levels in the normal fibroblast cells. We demonstrate that the 84BR and 175BR cells display a 2.06- and 2.09-fold increase in nuclear artemis expression. These data support our observations of elevated artemis expression derived from the Q-PCR experiments.
Artemis expression in the NB1-Tert parental cells was derived as explained above. Artemis expression in the vector-only-transfected cells and in clone 6 reveals a largely unchanged level of artemis expression. However, in clones 1–5, there is an increased level of artemis expression in the nuclei of these cells. This ranged from a 1.65-fold increased level of expression in clone 3 to a 2.25-fold elevated expression in clone 5. Representative images of artemis staining of cells derived from imaging flow cytometry is shown in Figure 7B.
Discussion
In this report, we have identified cellular radiosensitivity in two human fibroblast cell lines. The cell line 84BR was derived from a clinically radiosensitive breast cancer patient, whereas the 175BR cell line was derived from a cancer patient with multiple tumours of independent histological origin. We demonstrated that the cells were 1.5–2 fold more sensitive to the lethal effects of IR than the two fibroblast cell lines 1BR.3 and NB1 derived from repair normal individuals, using a clonogenic assay. We also demonstrated that the enhanced radiosensitivity in these cell lines was associated with a failure to effectively repair DNA DSB following radiation exposure as measured in a γ-H2AX foci retention assay. We also observed elevated levels of apoptosis prior to and after 2 Gy gamma radiation exposure in the two radiation-sensitive fibroblast cell lines. The Q-PCR analysis of key genes associated with NHEJ DNA repair revealed a 1.5–2-fold overexpression of the artemis endonuclease, a key component of the NHEJ-repair pathway. Moreover, we induced radiosensitivity and elevated apoptosis in an immortalised normal fibroblast cell line (NB1-Tert) by overexpression of artemis following transfection with the cDNA.
Defects in artemis are associated with human disease and a deficiency of artemis results in SCID-A, characterised by a loss and T- and B-cell function, elevated radiation sensitivity and increased cancer incidence (Moshous et al, 2000). In these cases, the phenotypes are usually associated with a reduction in the activity of artemis, leading to a failure of efficient NHEJ. However, we have demonstrated the corollary of this, in which cellular radiosensitivity appears to be associated with increased artemis expression. This seemingly counterintuitive observation is, notwithstanding, concordant with the recent report from Multhaup et al, 2010. Here the authors developed a series of lentiviral vectors containing the human artemis cDNA, which were used to transduce NIH3T3 cells. Overexpression of the artemis protein in these cells was associated with reduced cell survival, increased DNA damage and elevated apoptosis. Moreover, the overexpression of artemis was correlated with increased DNA DSB measured by comet assay. The authors concluded that the increased endonuclease activity attributable to the abnormally high cellular levels of the artemis protein was responsible for the reduced cell viability. Such observations have significant implications for gene-therapy approaches for the treatment of SCID. Gene therapy vectors resulting in overexpression of artemis are likely to be detrimental to genome integrity and may exacerbate the clinical symptoms of SCID. The overexpression of artemis in the 84BR and 175BR cell lines is also associated with reduced cell survival before and after exposure to gamma radiation. We further support these observations by the induction of cellular radiosensitivity in a normal fibroblast cell line NB1-Tert.
Although we demonstrate radiosensitivity associated with artemis overexpression in the two cell lines and that this result is concordant with that of Multhaup et al (2010), others have not demonstrated similar findings. For example, overexpression of functional artemis in the 48BR normal fibroblast cell line results in increased radioresistance. However, an artemis construct deleted at the C terminus thus removing endonuclease activity of the protein results in a dominant-negative phenotype, whereby the 48BR fibroblasts are rendered sensitive to the lethal effects of radiation and radiomimetic drugs (Mohapatra et al, 2011). The apparent disparity of these findings is unclear, but it may be speculated that cell line-specific differences in DNA repair and gene expression may underpin the differences in radiation survival following artemis transfection. Alternatively, overexpression of an endonuclease, such as artemis, beyond a critical level may lead to cytotoxicty rather than resistance to radiation.
Our findings together with the results of others (Mohapatra et al, 2011) indicate that both increased and reduced levels of artemis expression can result in cellular radiosensitivity. Although the level of overexpression in the 84BR and 175BR cell lines is approximately 1.5–2 fold, we do observe elevated apoptosis and a failure to efficiently repair DNA DSB before and following radiation exposure.
We hypothesise that in the cell lines described in this study, the increased expression of the artemis protein appears to act in a dominant-negative manner and can result in elevated sensitivity to IR and elevated apoptosis.
References
Abbaszadeh F, Clingen PH, Arlett CF, Plowman PN, Bourton EC, Themis M, Makarov EM, Newbold RF, Green MH, Parris CN (2010) A novel splice variant of the DNA-PKcs gene is associated with clinical and cellular radiosensitivity in a patient with xeroderma pigmentosum. J Med Genet 47 (3): 176–181
Arlett CF, Cole J, Green MHL (1989) Radiosensitive individuals in the population. In Low Dose Radiation: Biological Bases of Risk Assessment, Baverstock KF, Stather JW (eds) pp 240–252. Taylor Francis: London
Arlett CF, Green MH, Priestley A, Harcourt SA, Mayne LV (1988) Comparative human cellular radiosensitivity: I. The effect of SV40 transformation and immortalisation on the gamma-irradiation survival of skin derived fibroblasts from normal individuals and from ataxia-telangiectasia patients and heterozygotes. Int J Radiat Biol 54 (6): 911–928
Arlett CF, Plowman PN, Rogers PB, Parris CN, Abbaszadeh F, Green MH, McMillan TJ, Bush C, Foray N (2006) Clinical and cellular ionizing radiation sensitivity in a patient with xeroderma pigmentosum. Br J Radiol 79: 510–517
Bassing CH, Alt FW (2004) The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst) 3 (8-9): 781–796
Bourton EC, Plowman PN, Adam-Zahir S, Ulus-Senologulu G, Serrai H, Bottley G, Parris CN (2011) Multispectral imaging flow cytometry reveals distinct frequencies of γ-H2AX foci induction in DNA double strand break repair defective human cells lines. J Cyt A 81 (2): 130–137
Bridger JM, Kill IR (2004) Aging of Hutchinson–Gilford progeria syndrome fibroblasts is characterised by hyperproliferation and increased apoptosis. Exp Gerontol 39: 717–724
Burma S, Chen BP, Chen DJ (2006) Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 5: 1042–1048
Clingen PH, Arlett CF, Hartley JA, Parris CN (2007) Chemosensitivity of primary human fibroblasts with defective unhooking of DNA interstrand cross-links. Exp Cell Res 313: 753–760
Dvorak CC, Cowan MJ (2010) Radiosensitive severe combined immunodeficiency disease. Immunol Allergy Clin North Am 30 (1): 125–142
Hahn WC, Counter CM, Lundberg AS, Beijersberge RL, Brooks MW, Weinberg RA (1999) Creation of human tumour cells with defined genetic elements. Nature 400 (6743): 464–468
Ip SCY, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC (2008) Identification of Holliday junction resolvases from humans and yeast. Nature 456 (7220): 357–361
Kryston TB, Georgiev AB, Pissis P, Georgakilas AG (2011) Role of oxidative stress and DNA damage in human carcinogenesis. Mutat Res 711: 193–201
Livak KJ, Schmitten TD (2001) Analysis of gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408
Mohapatra S, Kawahara M, Khan IS, Yannone SM, Povirk LF (2011) Restoration of G1 chemo/radioresistance and double-strand-break repair proficiency by wild-type but not endonuclease-deficient artemis. Nucleic Acids Res 39 (15): 6500–6510
Moshous D, Li L, Chasseval R, Philippe N, Jabado N, Cowan MJ, Fischer A, de Villartay JP (2000) A new gene involved in DNA double-strand break repair and V(D)J recombination is located on human chromosome 10p. Hum Mol Genet 9: 583–588
Multhaup M, Karlen AD, Swanson DL, Wilber A, Sornia NV, Cowan MJ, McIvor RS (2010) Cytotoxicity associated with artemis overexpression after lentiviral vector-mediated gene transfer. Hum Gene Ther 21: 865–875
Musio A, Marrella V, Sobacchi C, Rucci F, Fariselli L, Giliani S, Lanzi G, Notarangelo LD, Delia D, Colombo R, Vezzoni P, Villa A (2005) Damaging-agent sensitivity of Artemis-deficient cell lines. Eur J Immunol 35 (4): 1250–1256
Ramsden DA, Weed BD, Reddy YVR (2010) V(D)J recombination: born to be wild. Semin Cancer Biol 20: 254–260
Riballo E, Critchlow SE, Teo SH, Doherty AJ, Priestley A, Broughton B, Kysela B, Beamish H, Plowman N, Arlett CF, Lehmann AR, Jackson SP, Jeggo PA (1999) Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr Biol 9 (13): 699–702
Rothkamm K, Krüger I, Thompson LH, Löbrich M (2003) Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol 23 (16): 5706–5715
Scott SP, Pandita TK (2006) The cellular control of DNA double strand breaks. J Cell Biochem 99: 1463–1475
van der Burg M, Ijspeert H, Verkaik NS, Turul T, Wiegant WW, Morotomi-Yano K, Mari PO, Tezcan I, Chen DJ, Zdzienicka MZ, van Dongen JJ, van Gent DC (2010) A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest 119 (1): 91–98
West RB, Yaneva M, Lieber MR (1998) Productive and nonproductive complexes of Ku and DNA-dependent protein kinase at DNA termini. Mol Cell Biol 18 (10): 5908–5920
Acknowledgements
We acknowledge technical assistance from Miss Christina Plowman. This work was supported in part by The Vidal Sassoon Foundation USA.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Ulus-Senguloglu, G., Arlett, C., Plowman, P. et al. Elevated expression of artemis in human fibroblast cells is associated with cellular radiosensitivity and increased apoptosis. Br J Cancer 107, 1506–1513 (2012). https://doi.org/10.1038/bjc.2012.443
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2012.443
Keywords
This article is cited by
-
Lymphopoiesis in transgenic mice over-expressing Artemis
Gene Therapy (2016)