Abstract
Background:
Lung adenocarcinoma (LADCA) patients with epidermal growth factor receptor (EGFR) mutations are in general associated with relatively high clinical response rate to EGFR-tyrosine kinase inhibitors (TKIs) but not all responded to TKI. It has therefore become important to identify the additional surrogate markers regarding EGFR-TKI sensitivity.
Methods:
We first examined the effects of EGFR-TKIs, gefitinib and erlotinib, upon cell proliferation of lung adenocarcinoma cell lines. We then evaluated the gene profiles related to EGFR-TKI sensitivity using a microarray analysis. Results of microarray analysis led us to focus on carcinoembryonic antigen-related cell adhesion molecule (CEACAM) family, CEACAM 3, 5, 6, 7, and 19, as potential further surrogate markers of EGFR-TKI sensitivity. We then examined the correlation between the status of CEACAM 3, 5, 6, 7, and 19 immunoreactivity in LADCA and clinicopathological parameters of individual cases.
Results:
In the cases with EGFR mutations, the status of all CEACAMs examined was significantly higher than that in EGFR wild-type patients, but there were no significant differences in the status of CEACAMs between TKI responder and nonresponder among 22 patients who received gefitinib therapy. However, among 115 EGFR mutation-negative LADCA patients, both CEACAM6 and CEACAM3 were significantly associated with adverse clinical outcome (CEACAM6) and better clinical outcome (CEACAM3).
Conclusion:
CEACAMs examined in this study could be related to the presence of EGFR mutation in adenocarcinoma cells but not represent the effective surrogate marker of EGFR-TKI in LADCA patients. However, immunohistochemical evaluation of CEACAM3/6 in LADCA patients could provide important information on their clinical outcome.
Similar content being viewed by others
Main
Epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor (TKI) has been reported to provide therapeutic benefits to NSCLC patients associated with EGFR gene mutations (Lynch et al, 2004; Paez et al, 2004) and also to female and nonsmoker patients (Thatcher et al, 2005). The response rate to EGFR-TKI among EGFR gene mutation-positive NSCLC patients has been reported as >70% and progression-free survival (PFS) as 9 to 10 months (Asahina et al, 2006; Inoue et al, 2006). Gefitinib did demonstrate a therapeutic effectiveness at least equivalent to docetaxel as the second-line chemotherapy in these patients with EGFR gene mutations (Niho et al, 2007). In addition, EGFR-TKI as the first-line therapy was reported to have extended the PFS of the EGFR mutation-positive lung cancer cases more significantly than the conventional chemotherapy (Mok et al, 2009; Maemondo et al, 2010). Erlotinib has also been reported to demonstrate a potential therapeutic benefit to the gefitinib-resistant EGFR mutation-positive lung cancer patients (Cho et al, 2007).
It has then become important to evaluate the potential surrogate markers of these EGFR-TKI agents in addition to the presence or absence of EGFR mutation(s) in order to increase the response rate to these agents. The first potential surrogate marker for primary resistance to EGFR-TKI reported in the literature was KRAS mutations in the EGFR mutations-negative cases (Shigematsu et al, 2005). Acquired clinical resistance to EGFR-TKI was also documented in lung cancer patients, who had an EGFR mutation in exon 20 (T790M) (Bell et al, 2005). In addition, the resistance to gefitinib and erlotinib in NSCLC cell lines was reported to be associated with epithelial-to-mesenchymal transition (EMT) of these cell lines examined (Thomson et al, 2005; Yauch et al, 2005; Witta et al, 2006). Therefore, in this study, we first examined the effects of gefitinib or erlotinib on cell proliferation of the cell lines including those originated from lung adenocarcinoma (LADCA). We then evaluated gene profiles of EGFR-TKI-sensitive cells using a microarray analysis in order to further characterise the possible differential mRNA expression patterns among EGFR-TKI-sensitive cells. These results of microarray analysis led us to focus on carcinoembryonic antigen-related cell adhesion molecule (CEACAM) family including carcinoembryonic antigen (CEA) as a potential surrogate marker of EGFR-TKI sensitivity. However, it is also true that the biological or clinical significance of CEACAM family expression including CEA in NSCLC has not necessarily been well characterised. Therefore, we also examined the relationship between the expression of CEACAM family and clinicopathological factors including patient outcome, EGFR mutation, and EGFR-TKI response in human LADCA cases in our present study.
Materials and methods
Cell lines
In this study, we used the following cell lines: A549, LCSC#1, RERF-LC-OK, LK87, and LCAM1. The original tissues, sources, and medium employed in these cell lines above are summarised in Supplementary Table S1. EGFR mutations in exons 18, 19, 20, and 21, which confer sensitivity to EGFR-TKI, were identified by the PCR-Invader assay (BML, Inc., Tokyo, Japan). Cells were maintained in each medium supplemented with 10% fetal bovine serum (Nichirei Co. Ltd, Tokyo, Japan). All the cells were maintained in culture at 37 °C, 95% relative humidity, and 5% CO2 at room air.
EGFR-TKI sensitivity test
Gefitinib was commercially obtained from Biaffin GmbH (Kassel, Germany). Erlotinib was kindly provided by Roche Diagnostics GmbH (Mannheim, Germany). Each cell lines above were cultured in a 96-well culture plate. At 72 h after gefitinib or erlotinib treatment, the cell number was evaluated using a Cell Counting Kit (DOJINDO LABORATORIES, Kumamoto, Japan) (Isobe et al, 1999). Then, 10 μl of 5 mM WST-8 was added to these cells, which were then incubated for 2 h at 37 °C. Optical densities (OD, 450 nm) were obtained with microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The status of cell proliferation (%) was calculated according to the following equation: (cell OD value after test materials treated/vehicle control cell OD value) × 100.
Microarray analysis
Cell lysates were prepared using RLT buffer (QIAGEN GmbH, Hilden, Germany). Total RNA was extracted using RNeasy Mini Kit (QIAGEN). First-strand cDNA was synthesised by incubating 5 μg of total RNA with 200 U SuperScript II reverse transcriptase (Invitrogen Corporation, Carlsbad, CA, USA) and 100 pmol T7-(dT)24 primer (Invitrogen). Ten units of T4 DNA polymerase (Invitrogen) were then added, and the dsDNA was mixed with T7 RNA polymerase (Invitrogen). The purified cRNA was fragmented at 300–500 bp as target solution. Both test and reference samples were labelled with cyanine-5 (Cy5)-labelled CTP (PerkinElmer Inc., Waltham, MA, USA). The Cy5-labelled cRNA probes were subsequently hybridised on the Human 1A version 2.0 (Agilent Technologies, Inc., Santa Clara, CA, USA) including 22 000 genes. The reacted arrays were then scanned as digital image files with GenePix 4000A (Axon Instruments, Foster City, CA, USA). Results were extracted using Agilent Feature Extraction software version 9.5.3.1 (Agilent Technologies) and analysed using Gene Spring GX 7.3.1 software (Agilent Technologies) in order to obtain gene expression ratios. Raw microarray data were normalised and analysed using the Gene Spring GX 7.3.1 software (Agilent Technologies). Expression data were median centred.
Patients and tissue specimens
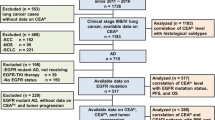
A total of 165 specimens of LADCA were obtained from the patients who underwent surgical resection from 2000 to 2006 in the Department of Surgery, Tohoku University Hospital and Miyagi Cancer Center. Clinicopathological features of the cases examined in this study are summarised in Supplementary Tables S2 and S3. A total of 115 cases were EGFR mutation-negative cases and had not received chemotherapy at all. Of the 165 LADCA patients, 50 were known to have EGFR mutations (exon 19 deletion, n=28; exon 21 point mutation, n=22; Supplementary Table S3). Among 50 LADCA cases, the response of gefitinib treatment was evaluated in 22 cases (responder (PR), n=15; nonresponder (SD), n=7; Supplementary Table S3). Time to progression was available in 17 out of these 22 cases who received gefitinib treatment, and hence 5 cases whom we lost afterward were treated as censored cases. Other EGFR mutation-positive 28 cases did not receive gefitinib treatment or no recurrence in their clinical course. All the specimens studied had been fixed in 10% formalin and embedded in paraffin wax. Research protocols for this study were approved by the Ethics Committee at Tohoku University School of Medicine (2009-380) and Miyagi Cancer Center (No. 34), respectively.
Immunohistochemistry
Primary antibodies used in this study were as follows: CEA/CEACAM5 (monoclonal CEM010; 1 : 1500 dilution; Mochida Pharmaceutical Co., Ltd, Tokyo, Japan), CEACAM6 (polyclonal, 1 : 200 dilution; Aviva Systems Biology, Corp., San Diego, CA, USA), CEACAM3 (polyclonal, 1 : 200 dilution; Sigma-Aldrich Corporation, St Louis, MO, USA), CEACAM7 (monoclonal BAC2, 1 : 200 dilution; Abcam plc, Cambridge, UK), and CEACAM19 (monoclonal HY-8H10, 1 : 300 dilution; Abcam). Streptavidin-biotin amplification method was employed for immunostaining using a Histofine Kit (Nichirei). The antigen–antibody complex was subsequently visualised with 3,3’-diaminobenzidine solution and counterstained with haematoxylin.
Evaluation of CEACAM immunohistochemistry was performed based on the staining proportion scoring systems used for CEACAM1 immunohistochemistry (Sienel et al, 2003; Dango et al, 2008) with some modifications. Immunoreactivity was examined independently by two of the authors (MK and YM) who were unaware of the clinical data. CEACAM immunoreactivity of tumour cells was compared with that in normal lung epithelial cells that were negative for immunoreactivity. CEACAM-positive rate was categorised according to the percentage of positive tumour cells into ‘negative’ (<40% positive carcinoma cells) and ‘positive’ (⩾40% positive carcinoma cells) (Sienel et al, 2003; Dango et al, 2008). Specificity of immunohistochemistry was assessed by evaluating the negative controls. For monoclonal antibodies, the primary antibodies had been replaced with normal rabbit nonimmune IgG. For polyclonal antibodies, immunoabsorption test using the corresponding antigens was conducted as a negative control.
Statistical analysis
The duration of disease-free survival (DFS) or PFS was calculated from the date of diagnosis to that of relapse or death, whichever first occurred, or to the last follow-up information for living patients (censored case). The duration of PFS was calculated from the date of start medication to that of progression, or to the last follow-up information for living patients (censored case). DFS and PFS data were graphically presented using the Kaplan–Meier method and were also compared with immunoreactivity of each CEACAM (positive vs negative) using the log-rank test. The 5-year DFS and PFS values were obtained from the Kaplan–Meier curves. The differences of positive rates of CEACAMs by each variant were assessed by Mann–Whitney U-test. The influence of each variable on the positive rate of each CEACAM was assessed by multinomial logistic regression model, and the survival of the patients was assessed by the Cox proportional hazards model. All statistical analyses were performed using Statview for windows (version 5.0; SAS Institute Inc., Cary, NC, USA). The accepted level of significance was P<0.05.
Results
EGFR-TKI sensitivity test
Results of the cell proliferation assays are summarised in Figure 1A. There was a significant decrease in the cell number after 48 h in RERF-LC-OK, A549, LCSC#1, and LK87 cells treated with 1 μ M (LCSC#1) or 10 μ M of erlotinib. There was a significant decrease in the cell number after 72 h in LCAM1, RERF-LC-OK, A549, LCSC#1, and LK87 cells treated with 1 μ M (LCSC#1 and LK87) or 10 μ M of gefitinib. There were no EGFR mutations in all these cell lines examined.
(A) Effects of EGFR-TKI on cell proliferation of the lung adenocarcinoma cell lines. Data are expressed as mean (n=3). (B) Each cell line was arranged according to the sensitivity of EGFR-TKI evaluated by EGFR-TKI sensitivity assay. We therefore searched gene expression similar to EGFR-TKI sensitivity patterns in five adenocarcinoma cell lines. In our present study, we focussed on four genes (CGM1 (CEACAM3), CD66c (CEACAM6), CGM2 (CEACAM7), and CEACAM19) of the CEACAM family.
The order of sensitivity to EGFR-TKI in the cells examined was as follows: LCSC#1, LK87, A549, RERF-LC-OK, and LCAM1.
Analysis of EGFR-TKI sensitivity-related genes using cDNA microarray
Each cell line was arranged according to the sensitivity of EGFR-TKI evaluated by EGFR-TKI sensitivity assay described above. We therefore searched gene expression similar to EGFR-TKI sensitivity patterns in five adenocarcinoma cell lines above (Figure 1B). In our present study, we focussed on four genes (CGM1 (CEACAM3), CD66c (CEACAM6), CGM2 (CEACAM7), and CEACAM19) of the CEACAM family.
CEACAMs in LADCA cases
We further examined the clinical significance of CEACAMs, especially that of CEA/CEACAM5, which has been known to be related to EGFR mutation (Okamoto et al, 2005; Shoji et al, 2007) in 115 EGFR mutation-negative LADCA cases using immunohistochemistry.
All CEACAMs examined were detected in cytoplasm and/or cell membrane of carcinoma cells (Figure 2). The positive cases of each CEACAM in 115 LADCA patients were summarised as follows: 36 cases (CEACAM5), 53 cases (CEACAM6), 57 cases (CEACAM3), 16 cases (CEACAM7), and 13 cases (CEACAM19). Results of an association between the status of CEACAM immunoreactivity and clinicopathological parameters in 115 LADCA patients are also summarised in Table 1. CEACAM3 status was significantly associated with gender or lymph node metastasis in 115 LADCA patients. There were no statistically significant association between other CEACAMs and clinicopathological parameters of individual patients.
The association between CEACAM expression and the 5-year DFS of the patients was evaluated using Kaplan–Meier survival curves and log-rank test. The statistical analysis demonstrated that the status of CEACAM3 in 115 LADCA patients examined in this study was significantly associated with better DFS, whereas that of CEACAM6 with poor DFS (Figure 3). The 5-year DFS rate of CEACAM6-negative patients was 74.2%, whereas that of CEACAM6-positive patients was 49.1%. Also, the 5-year DFS rate of CEACAM3-negative patients was 46.6%, whereas that of CEACAM3-positive patients was 78.9%. Using the Cox proportional hazards model, we performed multivariate analysis to assess the independent predictive value of CEACAM status for the DFS of patients with LADCA. The following prognostic variables were also included in this study: age, sex, tumour size, and lymph node metastasis. Results of this multivariate analysis demonstrated that CEACAM6 positive (P=0.0003), CEACAM3 positive (P=0.0008), age (P=0.032), tumour size (P=0.015), and lymph node metastasis (P=0.0001) were all turned out independent prognostic factors, respectively (Table 2).
Association between CEACAM status and EGFR mutation in LADCA cases
All CEACAM (CEA, CEACAM6, CEACAM7, CEACAM7, CEACAM19) immunoreactivity in EGFR mutation-positive cases was significantly higher than that in EGFR mutation-negative cases (Figure 4). However, there were no statistically significant differences in the status of CEACAMs between responder and nonresponder patients, and also in EGFR mutations between exon 19 and exon 21 among 22 EGFR mutation-positive LADCA patients (Figures 5 and 6). The association between CEACAM status and the PFS of the patients was evaluated using Kaplan–Meier survival curves and log-rank test. Results of univariate analysis demonstrated that the positive CEACAM6 status was associated with an increased PFS in EGFR mutation-positive LADCA patients with EGFR-TKI treatment (Figure 7). We also performed multivariate analysis, including age and gender, to assess the independent predictive value of CEACAM6 expression for PFS of the patients receiving EGFR-TKI treatment using Cox proportional hazards model but no significant correlations were detected (Supplementary Table S4).
Discussion
CEA is one of the most extensively studied tumour markers and belongs to the CEACAM family members. These groups of protein are typically cell membrane-associated glycoproteins, and are part of the immunoglobulin superfamily (Gold and Freedman, 1965). Among these CEACAM family, CEACAM5, also well known as CEA, was reported to be overexpressed in a majority of carcinomas including those of the gastrointestinal tract, the respiratory systems, and the breast (Hansen et al, 1974; Kuroki et al, 1992, Lamerz, 1999). CEACAM6 (CD66c, NCA-90) is a nonspecific crossreacting glycoprotein antigen that shares some antigenic determinants with CEACAM5 (Kuespert et al, 2006). CEACAM6 is also reported to be expressed in granulocytes and epithelia from various organs (Kuespert et al, 2006). Overexpression of CEACAM6 has been demonstrated to result in cell proliferation and invasion of breast and pancreatic cancer (Kuespert et al, 2006; Lewis-Wambi et al, 2008; Maraqa et al 2008). CEACAM3 is also present in neutrophils and considered to play an important role in the process of phagocytosis (Chen and Gotschlich, 1996). CEACAM7 expression was also very recently reported to be significantly low in rectal adenocarcinoma compared with that in normal mucosa (Messick et al, 2010). CEACAM19 has functional immunoreceptor tyrosine-based activation motifs in cytoplasmic domain (Kuespert et al, 2006), but it is also true that the physiological or pathological functions of CEACAM19 have remained entirely unknown at this juncture. CEACAMs have been also recently demonstrated to play important roles in several types of human malignancies, but the roles of CEACAMs have remained largely unknown in lung cancer.
In this study, we first demonstrated that the expression of CEACAM family (CEACAM 3, 6, 7, and 19) was associated with EGFR-TKI sensitivity in microarray analysis in vitro. The status of these CEACAMs was also significantly higher in EGFR mutation-positive cases than in negative LADCA cases. Shoji et al (2007) reported that serum CEA/CEACAM5 level was significantly higher in EGFR mutation-positive lung cancer cases than in wild-type cases. In addition, Okamoto et al (2005) demonstrated that in LADCA patients, serum CEA/CEACAM5 concentration of ⩾5 ng ml−1 turned out to be more sensitive to gefitinib treatment than those of ⩽5 ng ml−1. It is true that CEA/CEACAM5 was not included in EGFR-TKI sensitivity molecules examined by microarray analysis in our present study but CEA/CEACAM5 expression was significantly higher in EGFR mutation cases as well as other CEACAMs examined in our study compared with EGFR wild-type cases. There were, however, no significant statistical associations between the status of CEACAMs examined in primary tumour of the patients and clinical response of gefitinib treatment in 22 LADCA patients. Therefore, it awaits further investigations including the validation in a larger number of the cases in different institutions to clarify whether the status of these CEACAMs in adenocarcinoma cases actually results in EGFR TKI-sensitivity in LADCA patients or not.
In this study, we also examined the clinicopathological significance of CEACAMs in LADCA patients. Among 5 CEACAMs above, both CEACAM3 and CEACAM6 demonstrated the most significant clinical significance in terms of clinical outcome of the patients. Results of our present study clearly demonstrated that the positive rate of CEACAM3 was significantly higher in female or lymph node metastasis-negative LADCA patients. In addition, CEACAM3 and CEACAM6 positivity in carcinoma cells turned out to be independent prognostic factors in LADCA patients examined in this study, that is, CEACAM3 positivity was associated with significantly better prognosis and CEACAM6 positivity with significantly worse prognosis. CEACAM3 is well known to be present as transmembrane protein, whereas CEACAM6 is linked to membrane via glycosyl-phosphatidylinositol anchor in neutrophils (Kuespert et al, 2006). CEACAM6 also acts as an inducer of cell proliferation in A549 cells (Singer et al, 2010). A549 cells expressed significant amounts of nonmembrane-anchored variants of CEACAM6 as well as CEA/CEACAM5, representing a putative source for the increased CEACAM5/6 serum levels frequently detected in lung cancer patients (Singer et al, 2010). In our present study, CEACAM6-positive/CEACAM3-negative cases were significantly associated with poor clinical outcome compared with CEACAM6-negative/CEACAM3-positive, double-positive, and double-negative cases. CEACAM3, which is anchored in cell membrane, was also reported to form heterodimer with other CEACAM family including CEACAM6 (Skubitz and Skubitz, 2008). These findings, including results of our present study, all indicated that CEACAM3 may inhibit the dissociation of CEACAM6 from the cell membrane and the stimulatory effects upon cell proliferation of CEACAM6 in LADCA patients. In addition, CACAM3 status of the primary tumour turned out to be an independent factor of good prognosis in LADCA cases examined in this study. CEACAM3 may inhibit cell proliferation/invasion of LADCA cells as a binding protein, but further investigations are required for clarification. Results of the univariate analysis in our present study did demonstrate that CEACAM6 was associated with an increased PFS for EGFR mutation-positive adenocarcinoma patients undergoing EGFR-TKI treatment. EGFR gene mutation-positive lung cancer was also reported to demonstrate better treatment responses than EGFR mutation-negative lung cancers following EGFR-TKI treatment (Takano et al, 2008). These findings all suggest that CEACAM3 overexpression was associated with better prognosis or clinical outcome, and CEACAM6 overexpression could account for protecting EGFR-TKI resistance (Lo and Hung, 2006) in the EGFR mutation-positive LADCA patients. This is because CEACAM3- and CEACAM6-positive rates in EGFR mutation-positive cases were significantly higher than that in EGFR mutation-negative cases, and CEACAM6-positive cases receiving gefitinib therapy were associated with a relatively long PFS in their clinical course. In the present study, we did not get significant collection between CEACAM6 and the response to EGFR-TKI, but we found relatively high CEACAM6 expression in PR cases. Because we only dealt with SD or PR cases, or we did not have enough cases to assess, we thought of the possibility that there was no significant difference. We think it is necessary to assess the CEACAM6 expression of PD cases, T790M positive cases, and an independent larger set to confirm the assumption.
Abdel-Aziz et al (2009) reported that a double-positive status of CEA/CEACAM5 and EGFR expression was detected in the majority of patients (81%) with colorectal cancers. Abou-Rjaily et al (2004) also reported that CEACAM1 was closely associated with EGFR actions and may reduce the EGFR-mediated cell proliferation following EGF binding, and that the CEACAM1 effects upon EGF-dependent hepatocyte proliferation are mediated by its ability to bind to and sequester Shc, thus uncoupling EGFR signalling from the Ras/Raf/MAP kinase pathway (Abou-Rjaily et al, 2004). Therefore, the CEACAMs examined in our present study are reasonably postulated to be associated directly with EGFR and to modify the anti-tumour effects of EGFR-TKI in LADCA patients. Choi et al (2007) recently reported that CEACAM6 was decreased by gefitinib treatment and abundantly expressed in EGFR-mutant lung cancer cell lines. They also suggested that CEACAM6 could serve as a potentially important EGFR transcriptional target in these cell lines (Choi et al, 2007). Results of several previous studies also demonstrated the translocation of EGFR in the nucleus as full-length receptors (Marti et al, 1991; Lin et al, 2001; Li et al, 2009). Li et al (2009) demonstrated that expression of a nuclear localisation sequence-tagged EGFR in cetuximab-sensitive cells increased resistance to cetuximab, both in vitro and in mouse xenografts. These results as well as results of our own study all indicated that CEACAMs could interact with EGFR and subsequently stabilise EGFR on the cell membrane, and maintain the sensitivity of EGFR-TKI. Further investigations are, however, required to clarify further details of the biological correlations between CEACAMs and EGFR toward the development of much more effective EGFR-TKI therapy of NSCLC patients.
Statement of translational relevance
The CEACAMs (CEACAM 5, 3,6, 7, and 19) examined in this study could be effective surrogate markers for prediction of EGFR gene mutation. Among these five CEACAMs above, immunohistochemical evaluation of CEACAM3/6 in LADCA patients could contribute to predicting their clinical outcome.
Change history
26 October 2012
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Abdel-Aziz MM, Lotfy M, El-Kady IM, Abozaid M (2009) Mutant p53 protein in the serum of patients with colorectal cancer: correlation with the level of carcinoembryonic antigen and serum epidermal growth factor receptor. Cancer Detect Prev 32: 329–335
Abou-Rjaily GA, Lee SJ, May D, Al-Share QY, Deangelis AM, Ruch RJ, Neumaier M, Kalthoff H, Lin SH, Najjar SM (2004) CEACAM1 modulates epidermal growth factor receptor--mediated cell proliferation. J Clin Invest 114: 944–952
Asahina H, Yamazaki K, Kinoshita I, Sukoh N, Harada M, Yokouchi H, Ishida T, Ogura S, Kojima T, Okamoto Y, Fujita Y, Dosaka-Akita H, Isobe H, Nishimura M (2006) A phase II trial of gefitinib as firstline therapy for advanced non-small cell lung cancer with epidermal growth factor receptor mutations. Br J Cancer 95: 998–1004
Bell DW, Gore I, Okimoto RA, Godin-Heymann N, Sordella R, Mulloy R, Sharma SV, Brannigan BW, Mohapatra G, Settleman J, Haber DA (2005) Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 37: 1315–1316
Chen T, Gotschlich EC (1996) CGM1a antigen of neutrophils, a receptor of gonococcal opacity proteins. Proc Natl Acad Sci USA. 93: 14851–14856
Cho BC, Im CK, Park MS, Kim SK, Chang J, Park JP, Choi HJ, Kim YJ, Shin SJ, Sohn JH, Kim H, Kim JH (2007) Phase II study of erlotinib in advanced non–small-cell lung cancer after failure of gefitinib. J Clin Oncol 25: 2528–2533
Choi K, Creighton CJ, Stivers D, Fujimoto N, Kurie JM (2007) Transcriptional profiling of non-small cell lung cancer cells with activating EGFR somatic mutations. PLoS One 2: e1226
Dango S, Sienel W, Schreiber M, Stremmel C, Kirschbaum A, Pantel K, Passlick B (2008) Elevated expression of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM-1) is associated with increased angiogenic potential in non-small-cell lung cancer. Lung Cancer 60: 426–433
Gold P, Freedman SO (1965) Demonstration of tumor-specific antigens in human colonic carcinomata by immunological tolerance and absorption techniques. J Exp Med 121: 439–462
Hansen HJ, Snyder JJ, Miller E, Vandevoorde JP, Miller ON, Hines LR, Burns JJ (1974) Carcinoembryonic antigen (CEA) assay. A laboratory adjunct in the diagnosis and management of cancer. Hum Pathol 5: 139–147
Inoue A, Suzuki T, Fukuhara T, Maemondo M, Kimura Y, Morikawa N, Watanabe H, Saijo Y, Nukiwa T (2006) Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol 24: 3340–3346
Isobe I, Michikawa M, Yanagisawa K (1999) Enhancement of MTT, a tetrazolium salt, exocytosis by amyroid β-protein and chloroquine in cultured rat astrocytes. Neurosci Lett 266: 129–132
Kuespert K, Pils S, Hauck CR (2006) CEACAMs: their role in physiology and pathophysiology. Curr Opin Cell Biol 18: 565–571
Kuroki M, Matsuo Y, Kinugasa T, Matsuoka Y (1992) Three different NCA species, CGM6/CD67, NCA-95, and NCA-90, are comprised in the major 90 to 100-kDa band of granulocyte NCA detectable upon SDS-polyacrylamide gel electrophoresis. Biochem Biophys Res Commun 182: 501–506
Lamerz R (1999) Role of tumour markers, cytogenetics. Ann Oncol 10: 145–149
Lewis-Wambi JS, Cunliffe HE, Kim HR, Willis AL, Jordan VC (2008) Overexpression of CEACAM6 promotes migration and invasion of oestrogen-deprived breast cancer cells. Eur J Cancer 44: 1770–1779
Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL (2009) Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene 28: 3801–3813
Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC (2001) Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol 3: 802–808
Lo HW, Hung MC (2006) Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br J Cancer 94: 184–188
Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139
Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, Yoshimori K, Harada T, Ogura T, Ando M, Miyazawa H, Tanaka T, Saijo Y, Hagiwara K, Morita S, Nukiwa T North-East Japan Study Group (2010) Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N Engl J Med 362: 2380–2388
Maraqa L, Cummings M, Peter MB, Shaaban AM, Horgan K, Hanby AM, Speirs V (2008) Carcinoembryonic antigen cell adhesion molecule 6 predicts breast cancer recurrence following adjuvant tamoxifen. Clin Cancer Res 14: 405–411
Marti U, Burwen SJ, Wells A, Barker ME, Huling S, Feren AM, Jones AL (1991) Localization of epidermal growth factor receptor in hepatocyte nuclei. Hepatology 13: 15–20
Messick CA, Sanchez J, Dejulius KL, Hammel J, Ishwaran H, Kalady MF (2010) CEACAM-7: a predictive marker for rectal cancer recurrence. Surgery 147: 713–719
Niho S, Ichinose Y, Tamura T, Yamamoto N, Tsuboi M, Nakagawa K, Shinkai T, Jiang H, Nishiwaki Y, Fukuoka M (2007) Results of a randomized phase III study to compare the overall survival of gefitinib versus docetaxel in Japanese patients with non-small cell lung cancer who failed one or two chemotherapy regimens. J Clin Oncol 25: LBA7509 (2007 ASCO Annual Meeting Proceedings)
Okamoto T, Nakamura T, Ikeda J, Maruyama R, Shoji F, Miyake T, Wataya H, Ichinose Y (2005) The serum CEA level can be a predictive factor for the efficacy of gefitinib in NSCLC patients. Eur J Cancer 41: 1286–1290
Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500
Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, Fujisawa T, Feng Z, Roth JA, Herz J, Minna JD, Gazdar AF (2005) Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 97: 326–328
Shoji F, Yoshino I, Yano T, Kometani T, Ohba T, Kouso H, Takenaka T, Miura N, Okazaki H, Maehara Y (2007) Serum carcinoembryonic antigen level is associated with epidermal growth factor receptor mutations in recurrent lung adenocarcinomas. Cancer 110: 2793–2798
Sienel W, Dango S, Woelfle U, Morresi-Hauf A, Wagener C, Brümmer J, Mutschler W, Passlick B, Pantel K (2003) Elevated expression of carcinoembryonic antigen-related cell adhesion molecule 1 promotes progression of non-small cell lung cancer. Clin Cancer Res 9: 2260–2266
Singer BB, Scheffrahn I, Kammerer R, Suttorp N, Ergun S, Slevogt H (2010) Deregulation of the CEACAM expression pattern causes undifferentiated cell growth in human lung adenocarcinoma cells. PLoS One 5: e8747
Skubitz KM, Skubitz AP (2008) Interdependency of CEACAM-1, -3, -6, and -8 induced human neutrophil adhesion to endothelial cells. J Transl Med 6: 78
Takano T, Fukui T, Ohe Y, Tsuta K, Yamamoto S, Nokihara H, Yamamoto N, Sekine I, Kunitoh H, Furuta K, Tamura T (2008) EGFR mutations predict survival benefit from gefitinib in patients with advanced lung adenocarcinoma: a historical comparison of patients treated before and after gefitinib approval in Japan. J Clin Oncol 26: 5589–5595
Thatcher N, Chang A, Parikh P, Rodrigues Pereira J, Ciuleanu T, von Pawel J, Thongprasert S, Tan EH, Pemberton K, Archer V, Carroll K (2005) Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer (Iressa Survival Evaluation in Lung Cancer; ISEL). Lancet 366: 1527–1537
Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, Iwata KK, Gibson N, Haley JD (2005) Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res 65: 9455–9462
Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M (2009) Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361: 947–957
Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, Helfrich B, Dziadziuszko R, Chan DC, Sugita M, Chan Z, Baron A, Franklin W, Drabkin HA, Girard L, Gazdar AF, Minna JD, Bunn PA (2006) Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res 66: 944–950
Yauch RL, Januario T, Eberhard DA, Cavet G, Zhu W, Fu L, Pham TQ, Soriano R, Stinson J, Seshagiri S, Modrusan Z, Lin CY, O'Neill V, Amler LC (2005) Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res 11: 8686–8698
Acknowledgements
We appreciate Erina Iwabuchi and Katsuhio Ono (Department of Pathology, Tohoku University School of Medicine) for their skilful technical assistance in cell culture and immunohistochemistry despite enormous damages inflicted upon Tohoku University by the earthquake on 11 March 2012 which interrupted this study. The grant supports were as follows: Grant-in-Aid for Young Scientist (B) and Grant-in-Aid for Scientific Research (B), MEXT, Tokyo, Japan. YM and HS have received research funding from CHUGAI Pharmaceutical Company.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors (KM and HY-O) have ownership interest in CHUGAI Pharmaceutical Company, Shizuoka, Japan.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies the paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Kobayashi, M., Miki, Y., Ebina, M. et al. Carcinoembryonic antigen-related cell adhesion molecules as surrogate markers for EGFR inhibitor sensitivity in human lung adenocarcinoma. Br J Cancer 107, 1745–1753 (2012). https://doi.org/10.1038/bjc.2012.422
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2012.422
Keywords
This article is cited by
-
LINC02126 is a potential diagnostic, prognostic and immunotherapeutic target for lung adenocarcinoma
BMC Pulmonary Medicine (2022)
-
Therapeutic Effect of pHLIP-mediated CEACAM6 Gene Silencing in Lung Adenocarcinoma
Scientific Reports (2019)
-
Carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation
Oncogene (2018)
-
High expression of CEACAM19, a new member of carcinoembryonic antigen gene family, in patients with breast cancer
Clinical and Experimental Medicine (2017)
-
A549 cells adapted to high nitric oxide show reduced surface CEACAM expression and altered adhesion and migration properties
Tumor Biology (2015)