Abstract
Aim:
There is a high incidence of the antiplatelet drug clopidogrel resistance (CR) in Asian populations. Because clopidogrel is a prodrug, polymorphisms of genes encoding the enzymes involved in its biotransformation may be the primary influential factors. The goal of this study was to investigate the associations of polymorphisms of CYP3A4, NR1I2, CYP2C19 and P2RY12 genes with CR in Chinese patients with ischemic stroke.
Methods:
A total of 191 patients with ischemic stroke were enrolled. The patients were treated with clopidogrel for at least 5 days. Platelet function was measured by light transmission aggregometry. The SNPs NR1I2 (rs13059232), CYP3A4*1G (rs2242480), CYP2C19*2 (rs4244285) and P2RY12 (rs2046934) were genotyped.
Results:
The CR rate in this population was 36%. The CYP2C19*2 variant was a risk factor for CR (*2/*2+wt/*2 vs wt/wt, OR: 2.366, 95% CI: 1.180–4.741, P=0.014), whereas the CYP3A4*1G variant had a protective effect on CR (*1/*1 vs *1G/*1G+*1/*1G, OR: 2.360, 95% CI: 1.247–4.468, P=0.008). The NR1I2 (rs13059232) polymorphism was moderately associated with CR (CC vs TT+TC, OR: 0.533, 95% CI: 0.286–0.991, P=0.046). The C allele in P2RY12 (rs2046934) was predicted to be a protective factor for CR (CC+TC vs TT, OR: 0.407, 95% CI: 0.191–0.867, P=0.018). In addition, an association was found between hypertension and CR (P=0.022).
Conclusion:
The individuals with both the CYP2C19*2 allele and hypertension are at high risk of CR during anti-thrombosis therapy. The CYP3A4*1G allele, P2RY12 (rs2046934) C allele and NR1I2 (rs13059232) CC genotype may be protective factors for CR. The associated SNPs studied may be useful to predict clopidogrel resistance in Chinese patients with ischemic stroke.
Similar content being viewed by others
Introduction
Clopidogrel (Plavix®) is an irreversible P2Y12 receptor antagonist prescribed for the treatment of arteriosclerotic events in patients with recent stroke, myocardial infarction (MI) or established peripheral arterial disease1,2. Although clopidogrel is safe and effective in most patients, there is an inevitable variability in responsiveness among individuals3. Current investigations indicate that 4% to 30% of patients fail to attain platelet inhibition after clopidogrel therapy4,5,6, and this phenomenon is called clopidogrel resistance (CR). In fact, several clinical studies conducted in Chinese, Japanese and Korean patients reveal that the frequencies of clopidogrel resistance in Asians may vary from 20% to 65%, which highly exceeds the incidence reported in other ethnic groups7,8. The underlying mechanisms of clopidogrel resistance remain unknown. Previous studies indicate that genetic factors may greatly contribute to the variability of platelet activity8.
Because clopidogrel is a prodrug that requires a two-step oxidation process for activation in the liver, polymorphisms of genes encoding the metabolic enzymes involved in clopidogrel biotransformation may be the primary influential factors of clopidogrel responsiveness. CYP2C19 plays a substantial role in both of these steps by producing nearly 50% of the active CYP2C19 metabolites9. CYP3A4 also participates in hepatic clopidogrel metabolism, producing 39.8% of the active metabolites during the second oxidation step9. Pregnane X receptor (PXR), a member of the nuclear receptor subfamily 1 (NR1I2), is a transcriptional regulator of several metabolic enzymes, including CYP3A4 and CYP2C1910. It has been previously reported that the genetic polymorphisms in NR1I2 may account for the interindividual variation in drug metabolism and disposition in certain diseases11,12. In addition, the P2RY12 gene encodes the P2Y12 ADP receptor, and genetic variants of P2RY12 have been associated with alterations in the platelet inhibition response in patients13. Many studies have focused on the association between genetic polymorphisms and clopidogrel resistance14,15,16.
According to the clopidogrel black box warning released by the US FDA, CYP2C19 genotyping now allows clinicians to adjust the treatment therapy for an individual patient. The different CYP2C19 genotypes have a significant impact on the response of clopidogrel and the prognosis of patients with ischemic stroke17. Although the hepatic CYP2C19 LOF genotypes are associated with a lower clopidogrel responsiveness18,19, only 12% of the platelet response to clopidogrel can be explained by the presence of the CYP2C19*2 polymorphism, as suggested by an intervention (PAPI) study20,21. Thus, other potential genetic factors and mechanisms remain to be discovered, which together may contribute to the individual variability of clopidogrel efficacy22. Moreover, limited documents have been published concerning the relationship between genetic polymorphisms and clopidogrel resistance in Chinese ischemic stroke patients. The aim of this study was to investigate the association between the CYP2C19*2 (rs4244285), CYP3A4*1G (rs2242480), NR1I2 (rs13059232) and P2RY12 (rs2046934) polymorphisms and clopidogrel resistance in Chinese ischemic stroke patients.
Material and methods
Study population
This study was approved by the Ethics Committee of Guangdong Provincial Hospital of Traditional Chinese Medicine, China. It was conducted in accordance with the Declaration of Helsinki and was consistent with applicable guidelines for good clinical practice. In total, 191 ischemic stroke patients between 40 and 80 years of age were enrolled in the study from January 2014 to January 2015. The inclusion criteria were as follows: (1) clinical diagnosis of ischemic stroke according to the revised guideline of the 4th Cerebrovascular Disease Forum of China, as shown by CT or MRI; (2) treatment with clopidogrel for at least 5 d with complete laboratory examinations and clinical records; and (3) signed informed consent prior to the study. The exclusion criteria were as follows: (1) patients who were allergic to clopidogrel; (2) a platelet count greater than 450×109/L or less than 150×109/L; (3) anti-coagulation therapy with warfarin or heparin within 1 month of the study; (4) recent history of active bleeding; (5) ALT, AST or TB values 3-fold higher than the normal range; (6) serum creatinine 50% above the normal upper limit; and (7) major surgery within 1 month of the study. Depending on their condition, enrolled patients were treated with either a 300 mg loading dose of clopidogrel followed by a 75 mg maintenance dose per day or a 75 mg maintenance dose daily. Demographic characteristics, clinical information and medications were collected from patient electronic medical records at the Guangdong Provincial Hospital of Traditional Chinese Medicine.
Extraction of peripheral blood DNA
Whole blood samples were obtained from participants and collected in EDTA tubes and stored at −80 °C. The extraction method was performed based on a previous study with some modifications23. The concentration and purity of extracted DNA samples were calculated using a Nanodrop 2000 Spectrophotometer (Thermofisher, USA).
Genotype analysis
Four single nucleotide polymorphisms were selected for the study, including NR1I2 (rs13059232), CYP3A4*1G (rs2242480), CYP2C19*2 (rs4244285) and P2RY12 (rs2046934), and they were genotyped using Sequenom MassARRAY iPLEX technology (Sequenom, San Diego, CA, USA). Operations were based on the instructions for multiplexed genotyping analysis using iPLEX gold in a 96-well format for MassARRAY. DNA (10 ng) samples were amplified by multiplex PCRs, and the amplification products were subsequently used for iPLEX extension reactions. The final products were desalted and dispensed into a 96-pad SpectroCHIP. Allele detection was performed using MALDI-TOF mass spectrometry (Sequenom). MassARRAY Typer software was utilized to analyze the mass spectrums.
Platelet function assay
Platelet function was measured by the light transmission aggregometry method to evaluate antiplatelet responses. Before the next dose of clopidogrel was administered, peripheral venous blood samples were collected from patients. The platelet aggregation rate was measured within 3 h. The blood was centrifuged to separate the platelet-rich plasma (PRP) and the platelet-poor plasma (PPP), and the PRP sample was diluted to 2×109 to 3×109/L using the PPP sample. Next, both the PPP and PRP samples were heated to 37 °C for 3 min. To measure platelet aggregation, 10 μmol/L of ADP was added to the PRP sample to induce platelet aggregation, and the change in light transmission in the PRP sample was recorded until the response reached a plateau. The maximal platelet aggregation rate (MPAR) was calculated and recorded. After the baseline platelet aggregation measurements were obtained, follow-up platelet aggregation studies were repeated 5 d after the last dose of clopidogrel.
Definition of clopidogrel resistance
Clopidogrel resistance was defined as either less than a 10% change in MPAR on d 5 or the MPAR (d 5) was greater than 50% of the baseline magnitude (d 0)24,25. The platelet inhibition rate was quantified as follows:
Platelet inhibition rate (PIR)=PAR (d 0)–PAR (d 5).
PIR<10%×PAR (d 0) or PAR (d 5)>50% PAR (d 0) was defined as clopidogrel resistance. According to this standard, participants were divided into clopidogrel resistance (CR) or non-resistance (NCR) groups.
Statistical analysis
Data are presented as the mean±SD for continuous variables and as frequencies for categorical variables. The significant differences between groups were assessed using the Chi-square test (when variables were categorical) or the independent-samples t-test (when variables were continuous). Hardy-Weinberg equilibrium and allele frequency comparisons were predicted using the Chi-square test. A significance level of P<0.05 was based on the two-sided probability test. All statistical analysis was performed using SPSS version 21.0.
Results
Baseline characteristics of study participants
In total, 191 ischemic stroke patients with different disease statuses were enrolled. There were 110 males (58%) and 81 females (42%) included in our study. The average age of the participants was 67 years old. All of the patients were divided into the NCR or CR group according to the definitions specified in the methods section. The CR rate was 36% of the research population. In our study population, 131 subjects were in an acute phase, 11 subjects were in a recovery phase, 32 subjects were in a sequela phase and 17 subjects were suffering from transient ischemia. No discrepancies in the CR rate were found among these four categories of stroke patients. Other characteristics of the study population are presented in Table 1. No significant differences in the distribution of age, sex, diabetes, smoking, alcohol, medications, previous history, NIHSS score or biochemical indexes were found between the CR and NCR groups. Patients with hypertension exhibited a higher prevalence of CR (81% with hypertension in 69 patients).
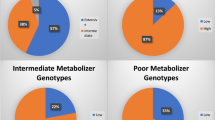
Genotype distributions and allele frequencies of CYP2C19*2, CYP3A4*1G, NR1I2 and P2RY12
Table 2 shows the genotype distributions and allele frequencies of the four selected SNPs. Allele frequencies of all SNPs were in Hardy-Weinberg equilibrium (P>0.05). The frequencies of the mutant alleles and genotypes of CYP3A4*1G, CYP2C19*2 and P2RY12 (rs2046934) were similar to previously reported findings13,15,26. The genotype distribution and allele frequency of NR1I2 (rs13059232) was not documented well in research articles; therefore, we consulted the NCBI database and found that there was no significant difference between the results in the database and our data.
Association of CYP2C19, CYP3A4, NR1I2 and P2RY12 gene polymorphisms with clopidogrel resistance
The association of CYP2C19, CYP3A4, NR1I2 and P2RY12 gene polymorphisms with clopidogrel resistance are shown in Table 3. CYP3A4*1G was significantly associated with a lower rate of clopidogrel resistance (*1/*1 vs *1G/*1G+*1/*1G, OR: 2.360, 95% CI: 1.247–4.468, P=0.008), and the CR rate was higher in the *1/*1 genotype than in those carrying the variant genotypes *1G/*1G and *1/*1G (25.1% vs 14.3%). NR1I2 rs13059232 was moderately associated with CR (CC vs TT+TC, OR: 0.533, 95% CI: 0.286–0.991, P=0.046). Subjects with mutant homozygotes (CC) of NR1I2 rs13059232 had a lower rate of CR than those carrying the T allele (16.1% vs 21.0%). CYP2C19*2 allele carriers had a higher risk of CR compared with wt/wt carriers (21.1% vs 13.6%). P2RY12 rs2046934 was positively associated with CR (CC+TC vs TT, OR: 0.407, 95% CI: 0.191–0.867, P=0.018), and mutant allele (C) carriers exhibited a lower rate of CR compared with wild-type homozygotes (TT) (6.2% vs 29.4%).
Discussion
The prevalence of clopidogrel resistance in our study population was approximately 36% (n=69), which is within the previously reported range7,8. Previous studies have shown that the antiplatelet effect of clopidogrel is achieved after taking a daily dose of 75 mg for 3 to 7 d4,27. Michelson et al reported that clopidogrel resistance may be explained by a pre-existing difference in the platelet response to ADP, indicating that there may be interindividual variation in ADP-induced platelet aggregation at baseline before clopidogrel therapy28. Therefore, we compared the platelet aggregation rate on d 0 with the follow-up rate on d 5 (defined as the steady state of clopidogrel treatment) in each patient. No variation in MPAR (d 0) was detected between the CR and NCR groups, and we observed significantly decreased platelet activity on d 5.
CYP2C19*2 was an independent predictor of clopidogrel resistance in patients with cardiovascular and cerebrovascular diseases. In previous studies15,17, the CYP2C19*2 variant was widely accepted as a risk factor for clopidogrel resistance, and this was confirmed in our study (*2/*2+wt/*2 vs wt/wt, OR: 2.366, 95% CI: 1.180–4.741, P=0.014). The CR rate of subjects with the *2/*2 and wt/*2 genotypes was 7.6% higher than those with the wt/wt genotype, and the estimated CR risk of the CYP2C19*2 allele carriers was 2.366-fold higher than in the wt/wt carriers. Therefore, these results confirm that the CYP2C19*2 allele reduced the activity of CYP2C19, decreasing the production of active metabolites and thereby impairing clopidogrel-induced platelet inhibition.
CYP3A4*1G, a SNP in intron 10 of CYP3A4 and characterized by a G to A substitution at position 82266, was the most frequent mutant allele of CYP3A4 in Asians. The allele frequency of CYP3A4*1G was 0.22-0.37 in the Chinese population26,29. This mutant genotype was correlated with a higher CYP3A metabolic activity, thus increasing the formation of metabolites, particularly for prodrugs, which are activated through biotransformation via CYP3A30. In addition, there is evidence of an association between the presence of CYP3A4*1G and clopidogrel response variability in Spanish patients with stable coronary artery disease31. However, the relationship between CYP3A4*1G and clopidogrel resistance in Chinese patients with ischemic stroke had not been investigated previously. In this study, we found that the CYP3A4*1G variant had a protective effect on the clopidogrel resistance of ischemic stroke patients in China. The CYP3A4*1/*1 genotype added a 10.8% increased risk of CR, which resulted in a 2.360-fold higher estimated risk for *1/*1 carriers of CR compared with *1G allele carriers. The variant allele increased the activity of CYP3A4, leading to higher production of active metabolites and thereby increasing the antiplatelet efficacy of clopidogrel. Therefore, the CYP3A4*1G variant was determined to be a protective factor in clopidogrel resistance.
It is well known that the Pregnane X Receptor (NR1I2), an upstream regulator of CYP450s, is closely associated with the metabolism of xenobiotics and endogenous substances. Recently, many SNPs in the NR1I2 promoter and intron regions were studied in different races32,33. NR1I2 (rs13059232), a SNP in intron 1 of NR1I2, was consistently associated with CYP3A4 phenotypic measures. In our investigation, NR1I2 (rs13059232) was moderately associated with clopidogrel resistance (P=0.046, OR: 0.533). The estimated CR risk of subjects carrying the T allele was 1.876-fold higher than those carrying the mutant genotype (CC). Because NR1I2 (rs13059232) may influence the phenotype of CYP3A434,35 and the CYP3A4*1G polymorphism would affect the enzyme activity of CYP3A4, there is potential interaction between CYP3A4*1G and NR1I2 (rs13059232). In this study, we found a moderate association between NR1I2 (rs13059232) and CR in Chinese ischemic stroke patients, which had not been previously reported.
The P2RY12 gene encodes the P2Y12 receptor protein, which is the pharmacological target of clopidogrel. Genetic polymorphisms of the P2Y12 ADP receptor might alter the receptor activation induced by ADP or the response to platelet inhibitors in patients36. Goran et al reported that P2RY12 (rs2046934) was consistently associated with a higher platelet reactivity in patients on clopidogrel in the ADP-induced LTA and the VerifyNow P2Y12 assay; this may provide protection against atherothrombotic events16. Robert et al provided evidence for an association between the P2RY12 haplotype H2 (composed of dbSNP rs10935838, rs2046934, rs5853517 and rs6809699) and a lower risk of DVT/PE37. We found that the P2RY12 polymorphism was significantly associated with individual variation of clopidogrel sensitivity in ischemic stroke patients. In the current study, P2RY12 (rs2046934) mutant allele carriers (CC+TC) exhibited a lower risk of CR than TT carriers, with an OR of 0.407. Therefore, P2RY12 (rs2046934) was included as one of the variations in the P2RY12 gene that was examined and was found to contribute to interindividual variability during clopidogrel therapy. The P2RY12 (rs2046934) C allele also exhibited a protective effect on CR.
In the current study, hypertension was found as a non-genetic factor significantly associated with the antiplatelet effects of clopidogrel. A correlation between CR and hypertension was also documented in a recent study conducted on 303 ischemic cerebral infarction patients in China38. However, the relationship between hypertension and the incidence of clopidogrel resistance remains unclear. Although diabetes mellitus was previously suggested to influence the antiplatelet effects of clopidogrel39,40, we did not observe any correlation between CR and diabetes mellitus in our study population. In addition, sex, age, smoking, drinking or other biomedical indexes were not associated with CR in ischemic stroke patients. Concomitant use of lipophilic statins (atorvastatin, simvastatin) and calcium-channel blockers were suspected to be associated with a diminished antiplatelet response, which is likely due to a shared metabolic pathway via the CYP2C19 or CYP3A4 isoenzymes41,42. Therefore, we evaluated the association between CR and the co-administration of statins, anti-hypertensive drugs and other co-administered drugs that might affect the efficacy of clopidogrel according to previous reports43,44. Our results did not demonstrate a significant association of co-medication with CR in our research population. Several previous investigations similar to ours also did not find any obvious association between CR and potential drug-drug interactions45,46,47. Therefore, combination therapy might have an inconsequential impact and likely does not contribute to the influence of genetic factors on CR. Discrepancies in non-genetic factors influencing CR occurrence may vary among population and races. In addition, the impact of non-genetic factors may be masked by the effect of genetic factors.
In summary, our findings indicate that the CYP2C19*2 allele and hypertension are risk factors for CR, whereas the CYP3A4*1G allele, P2RY12 (rs2046934) C allele and NR1I2 (rs13059232) CC genotype are protective against CR in Chinese ischemic stroke patients. Because our study was limited by the sample size, further investigations should be conducted to verify the associated genetic and non-genetic factors with CR. The current results may be useful for predicting the development of clopidogrel resistance in different patient populations.
Abbreviations
MI, myocardial infarction; PAPI, pharmacogenomics of antiplatelet intervention; CT, computed tomography; MRI, magnetic resonance imaging; TB, total bilirubin; CR, clopidogrel resistance; NCR, non-clopidogrel resistance; OR, odds ratio; CI, confidence interval; MPAR, max platelet aggregation rate; EDTA, ethylenediaminetetraacetic acid; PD, pharmacodynamics; PK, pharmacokinetics; SNP, single nucleotide polymorphism; NIHSS, National Institutes of Health stroke scale; ADP, adenosine diphosphate; ALT, alanine transaminase; AST, aspartate aminotransferase; PLT, platelet; TC, total cholesterol; TG, triglyceride; LTA, light transmission aggregation; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; HWE, Hardy-Weinberg equilibrium; TIA, transient ischemic attack.
Author contribution
Jin JING, Min HUANG, and Rui LIU designed the study; Rui LIU, Zi-yi ZHOU, and Ye-feng CAI performed the research; Min ZHAO, Yuan-qi ZHAO, Wei-bang YU, and Yi-bei CHEN assisted with the research; Xin-meng CHEN contributed reagents; Rui LIU analyzed data; Rui LIU, Jia-li LI, and Jin JING wrote the paper.
References
Kelly RP, Close SL, Farid NA, Witers KJ, Shen L, Natanegara F, et al. Pharmacokinetics and pharmacodynamics following maintenance doses of prasugrel and clopidogrel in Chinese carriers of CYP2C19 variants. Br J Clin Pharmacol 2012; 73: 93–105.
Boggon R, van Staa TP, Timmis A, Hemingway H, Ray KK, Begg A, et al. Clopidogrel discontinuation after acute coronary syndromes: frequency, predictors and associations with death and myocardial infarction — a hospital registry - primary care linked cohort (MINAP-GPRD). Eur Heart J 2011; 32: 2376–86.
Perry CG, Shuldiner AR . Pharmacogenomics of anti-platelet therapy: how much evidence is enough for clinical implementation? J Hum Genet 2013; 58: 339–45.
Nguyen TA, Diodati JG, Pharand C . Resistance to clopidogrel: a review of the evidence. J Am Coll Cardiol 2005; 45: 1157–64.
Serebruany VL, Steinhubl SR, Berger PB, Malinin AI, Bhatt DL, Topol EJ . Variability in platelet responsiveness to clopidogrel among 544 individuals. J Am Coll Cardiol 2005; 45: 246–51.
Ford NF . Clopidogrel resistance: pharmacokinetic or pharmacogenetic? J Clin Pharmacol 2009; 49: 506–12.
Hasan MS, Basri HB, Hin LP, Stanslas J . Genetic polymorphisms and drug interactions leading to clopidogrel resistance: why the Asian population requires special attention. Int J Neurosci 2013; 123: 143–54.
Jiang XL, Samant S, Lesko LJ, Schmidt S . Clinical pharmacokinetics and pharmacodynamics of clopidogrel. Clin Pharmacokinet 2015; 54: 147–66.
Kazui M, Nishiya Y, Ishizuka T, Hagihara K, Farid NA, Okazaki O, et al. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos 2010; 38: 92–9.
Wang XD, Deng XY, Chen J, Li JL, Chen X, Zhao LZ, et al. Single nucleotide polymorphisms of the pregnane X receptor gene in Han Chinese and a comparison with other ethnic populations. Pharmacology 2008; 81: 350–4.
Castano G, Burgueno A, Fernandez Gianotti T, Pirola CJ, Sookoian S . The influence of common gene variants of the xenobiotic receptor (PXR) in genetic susceptibility to intrahepatic cholestasis of pregnancy. Aliment Pharmacol Ther 2010; 31: 583–92.
Zhang B, Xie W, Krasowski MD . PXR: a xenobiotic receptor of diverse function implicated in pharmacogenetics. Pharmacogenomics 2008; 9: 1695–709.
Fontana P, Dupont A, Gandrille S, Bachelot-Loza C, Reny JL, Aiach M, et al. Adenosine diphosphate-induced platelet aggregation is associated with P2Y12 gene sequence variations in healthy subjects. Circulation 2003; 108: 989–95.
Frelinger AL 3rd, Bhatt DL, Lee RD, Mulford DJ, Wu J, Nudurupati S, et al. Clopidogrel pharmacokinetics and pharmacodynamics vary widely despite exclusion or control of polymorphisms (CYP2C19, ABCB1, PON1), noncompliance, diet, smoking, co-medications (including proton pump inhibitors), and pre-existent variability in platelet function. J Am Coll Cardiol 2013; 61: 872–9.
Tresukosol D, Suktitipat B, Hunnangkul S, Kamkaew R, Poldee S, Tassaneetrithep B, et al. Effects of cytochrome P450 2C19 and paraoxonase 1 polymorphisms on antiplatelet response to clopidogrel therapy in patients with coronary artery disease. PLoS One 2014; 9: e110188.
Rudez G, Bouman HJ, Van Werkum JW, Leebeek FW, Kruit A, Ruven HJ, et al. Common variation in the platelet receptor P2RY12 gene is associated with residual on-clopidogrel platelet reactivity in patients undergoing elective percutaneous coronary interventions. Circ Cardiovasc Genet 2009; 2: 515–21.
Jia DM, Chen ZB, Zhang MJ, Yang WJ, Jin JL, Xia YQ, et al. CYP2C19 polymorphisms and antiplatelet effects of clopidogrel in acute ischemic stroke in China. Stroke 2013; 44: 1717–9.
Hulot JS, Bura A, Villard E, Azizi M, Remones V, Goyenvalle C, et al. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood 2006; 108: 2244–7.
Liu Y, Liu N, Li W, Shao H, Zhi H, Li J . Relationship of CYP2C19*2 and CYP2C19*3 gene polymorphism with clopidogrel response variability and recurrent cardiovascular events in Chinese patients undergoing percutaneous coronary intervention. Pharmacology 2013; 91: 165–72.
Cuisset T, Morange PE, Alessi MC . Recent advances in the pharmacogenetics of clopidogrel. Hum Genet 2012; 131: 653–64.
Shuldiner AR, O'Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 2009; 302: 849–57.
Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Meneveau N, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med 2009; 360: 363–75.
Loparev VN, Cartas MA, Monken CE, Velpandi A, Srinivasan A . An efficient and simple method of DNA extraction from whole blood and cell lines to identify infectious agents. J Virol Methods 1991; 34: 105–12.
Bliden KP, DiChiara J, Tantry US, Bassi AK, Chaganti SK, Gurbel PA . Increased risk in patients with high platelet aggregation receiving chronic clopidogrel therapy undergoing percutaneous coronary intervention: is the current antiplatelet therapy adequate? J Am Coll Cardiol 2007; 49: 657–66.
Gurbel PA, Bliden KP, Hayes KM, Yoho JA, Herzog WR, Tantry US . The relation of dosing to clopidogrel responsiveness and the incidence of high post-treatment platelet aggregation in patients undergoing coronary stenting. J Am Coll Cardiol 2005; 45: 1392–6.
Du J, Xing Q, Xu L, Xu M, Shu A, Shi Y, et al. Systematic screening for polymorphisms in the CYP3A4 gene in the Chinese population. Pharmacogenomics 2006; 7: 831–41.
Savcic M, Hauert J, Bachmann F, Wyld PJ, Geudelin B, Cariou R . Clopidogrel loading dose regimens: kinetic profile of pharmacodynamic response in healthy subjects. Semin Thromb Hemost 1999; 25: 15–9.
Michelson AD, Linden MD, Furman MI, Li Y, Barnard MR, Fox ML, et al. Evidence that pre-existent variability in platelet response to ADP accounts for 'clopidogrel resistance'. J Thromb Haemost 2007; 5: 75–81.
Du J, Yu L, Wang L, Zhang A, Shu A, Xu L, et al. Differences in CYP3A41G genotype distribution and haplotypes of CYP3A4, CYP3A5 and CYP3A7 in 3 Chinese populations. Clin Chim Acta 2007; 383: 172–4.
Fukushima-Uesaka H, Saito Y, Watanabe H, Shiseki K, Saeki M, Nakamura T, et al. Haplotypes of CYP3A4 and their close linkage with CYP3A5 haplotypes in a Japanese population. Hum Mutat 2004; 23: 100.
Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, Ramirez C, Cavallari U, Trabetti E, et al. Contribution of gene sequence variations of the hepatic cytochrome P450 3A4 enzyme to variability in individual responsiveness to clopidogrel. Arterioscler Thromb Vasc Biol 2006; 26: 1895–900.
Choong E, Polari A, Kamdem RH, Gervasoni N, Spisla C, Jaquenoud Sirot E, et al. Pharmacogenetic study on risperidone long-acting injection: influence of cytochrome P450 2D6 and pregnane X receptor on risperidone exposure and drug-induced side-effects. J Clin Psychopharmacol 2013; 33: 289–98.
Chew SC, Lim JS, Lee EJ, Chowbay B . Genetic variations of NR2A1 in Asian populations: implications in pharmacogenetics studies. Drug Metab Pharmacokinet 2013; 28: 278–88.
Lamba J, Lamba V, Strom S, Venkataramanan R, Schuetz E . Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab Dispos 2008; 36: 169–81.
Schipani A, Siccardi M, D'Avolio A, Baietto L, Simiele M, Bonora S, et al. Population pharmacokinetic modeling of the association between 63396C→T pregnane X receptor polymorphism and unboosted atazanavir clearance. Antimicrob Agents Chemother 2010; 54: 5242–50.
Zoheir N, Abd Elhamid S, Abulata N, El Sobky M, Khafagy D, Mostafa A . P2Y12 receptor gene polymorphism and antiplatelet effect of clopidogrel in patients with coronary artery disease after coronary stenting. Blood Coagul Fibrinolysis 2013; 24: 525–31.
Zee RY, Michaud SE, Diehl KA, Chasman DI, Emmerich J, Gaussem P, et al. Purinergic receptor P2Y, G-protein coupled, 12 gene variants and risk of incident ischemic stroke, myocardial infarction, and venous thromboembolism. Atherosclerosis 2008; 197: 694–9.
Su JF, Hu XH, Li CY . Risk factors for clopidogrel resistance in patients with ischemic cerebral infarction and the correlation with gene rs1045642 polymorphism. Exp Ther Med 2015; 9: 267–71.
Iakovou I, Schmidt T, Bonizzoni E, Ge L, Sangiorgi GM, Stankovic G, et al. Incidence, predictors, and outcome of thrombosis after successful implantation of drug-eluting stents. JAMA 2005; 293: 2126–30.
Ang L, Palakodeti V, Khalid A, Tsimikas S, Idrees Z, Tran P, et al. Elevated plasma fibrinogen and diabetes mellitus are associated with lower inhibition of platelet reactivity with clopidogrel. J Am Coll Cardiol 2008; 52: 1052–9.
Tantry US, Jeong YH, Gurbel PA . The clopidogrel-statin interaction. Circ J 2014; 78: 592–4.
Tirkkonen T, Heikkilä P, Vahlberg T, Huupponen R, Laine K . Epidemiology of CYP3A-mediated clopidogrel drug-drug interactions and their clinical consequences. Cardiovasc Ther 2013; 31: 344–51.
Lau WC, Waskell LA, Watkins PB, Neer CJ, Horowitz K, Hopp AS, et al. Atorvastatin reduces the ability of clopidogrel to inhibit platelet aggregation a new drug-drug interaction. Circulation 2003; 107: 32–7.
Siller-Matula JM, Lang I, Christ G, Jilma B . Calcium-channel blockers reduce the antiplatelet effect of clopidogrel. J Am Coll Cardiol 2008; 52: 1557–63.
Saw J, Brennan DM, Steinhubl SR, Bhatt DL, Mak KH, Fox K, et al. Lack of evidence of a clopidogrel-statin interaction in the CHARISMA trial. J Am Coll Cardiol 2007; 50: 291–5.
Ojeifo O, Wiviott SD, Antman EM, Murphy SA, Udell JA, Bates ER, et al. Concomitant administration of clopidogrel with statins or calcium-channel blockers: Insights from the TRITON-TIMI 38 (Trialto Assess Improvement in Therapeutic Outcomes by Optimizing Platelet Inhibition With Prasugrel-Thrombolysis In MyocardialInfarction 38). JACC Cardiovasc Interv 2013; 6: 1275–81.
Pelliccia F, Rosano G, Marazzi G, Vitale C, Spoletini I, Franzoni F, et al. Pharmacodynamic effects of atorvastatin versus rosuvastatin in coronary artery disease patients with normal platelet reactivity while on dual antiplatelet therapy — The PEARL randomized cross-over study. Eur J Pharmacol 2014; 725: 18–22.
Acknowledgements
This work was supported by the National Major Projects for Science and Technology Development from the Science and Technology Ministry of China (Grant No 2012ZX09506001-004) and the Fundamental Research Funds for the Central Universities (No 16ykpy14). The authors thank the Guangdong Provincial Hospital of Traditional Chinese Medicine for their cooperation.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Liu, R., Zhou, Zy., Chen, Yb. et al. Associations of CYP3A4, NR1I2, CYP2C19 and P2RY12 polymorphisms with clopidogrel resistance in Chinese patients with ischemic stroke. Acta Pharmacol Sin 37, 882–888 (2016). https://doi.org/10.1038/aps.2016.41
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2016.41
Keywords
This article is cited by
-
The stent thrombosis in Belgium (STIB) scoring system reliability in Indonesia patients and the modified STIB scoring (M-STIB)
The Egyptian Journal of Neurology, Psychiatry and Neurosurgery (2023)
-
Association between P2Y1 and P2Y12 polymorphisms and acute myocardial infarction and ADP-induced platelet aggregation
BMC Cardiovascular Disorders (2023)
-
Baseline P2Y12 reactivity, kidney function, and CYP2C19 genotype determine clopidogrel responsiveness in acute stroke
Scientific Reports (2023)
-
Influence of Genetic and Epigenetic Factors of P2Y12 Receptor on the Safety and Efficacy of Antiplatelet Drugs
Cardiovascular Drugs and Therapy (2022)
-
Cardiovascular Pharmacogenomics: An Update on Clinical Studies of Antithrombotic Drugs in Brazilian Patients
Molecular Diagnosis & Therapy (2021)