Abstract
Nuclear receptors (NRs) are ligand-regulated transcription factors that regulate metabolism, development and immunity. The NR superfamily is one of the major classes of drug targets for human diseases. Retinoic acid receptor-related orphan receptor (ROR) α, β and γ belong to the NR superfamily, and these receptors are still considered as 'orphan' receptors because the identification of their endogenous ligands has been controversial. Recent studies have demonstrated that these receptors are regulated by synthetic ligands, thus emerge as important drug targets for the treatment of multiple sclerosis, rheumatoid arthritis, psoriasis, etc. Studying the structural basis and ligand development of RORs will pave the way for a better understanding of the roles of these receptors in human diseases. Here, we review the structural basis, disease relevance, strategies for ligand identification, and current status of development of therapeutic ligands for RORs.
Similar content being viewed by others
Introduction
The nuclear receptors (NRs) are a large family of ligand-regulated transcriptional factors and include the receptors for steroid hormones, thyroid hormones, lipophilic vitamins, and cholesterol metabolites1,2,3. NRs are involved in a wide variety of biological processes, such as cell proliferation, differentiation, development, and homeostasis4,5. Dysfunction of NR signaling leads to various diseases such as cancer, diabetes, obesity, and autoimmune disorders. The NR superfamily is one of the primary classes of therapeutic drug targets for human disease. The agonists or antagonists of NRs account for approximately 13% of all FDA-approved drugs6. Among the top-selling drugs, these include tamoxifen for estrogen receptors in breast cancer, dexamethasone for the glucocorticoid receptor in inflammatory diseases and rosiglitazone for the peroxisome proliferator-activated receptor-γ (PPARγ) in diabetes.
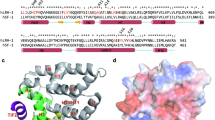
The NR superfamily is composed of 48 members in humans. The members share high sequence identity and conserved domains (Figure 1). The typical domain structure of NRs comprises four major functional regions (Figure 1A). The A/B region refers to the amino-terminal ligand-independent activation function 1 (AF-1) domain. The C region is the highly conserved DNA-binding domain (DBD) and contains two zinc finger motifs to bind distinct DNA response elements. The relatively short but flexible D (hinge) region links the C region to the E region. The E/F region is the carboxy-terminal ligand-binding domain (LBD) with the ligand-dependent activation domain 2 (AF-2). AF-1 and the hinge region are the most divergent, whereas the DBD and LBD regions are the most conserved in sequence and length across the superfamily. Ligand binding induces a conformational change of the LBD and alters the surface to facilitate the recruitment of cofactor proteins, which transcriptionally regulates the induction or repression of target genes (Figure 2).
Structural organization of ROR functional domains. (A) Schematic diagram of the domain structure of RORs. Similar to other NRs, RORs display conserved modular domain architecture with a N-terminal ligand-independent activation function 1 (AF-1) domain, followed by a DNA binding domain (DBD), a hinge domain, and a ligand-binding domain with an activation function 2 (AF-2) domain. The DBD binds specific DNA sequences that typically consist of TAAA/TNTAGGTCA (termed ROR response element, RORE). (B) Sequence alignment of the ligand binding domain of RORα, RORβ, and RORγ performed using ClustalW. Cartoon presentation of the general architecture of RORs was shown under the corresponding sequences. Identical residues are labeled with an asterisk. Partially conserved residues are labeled with a colon. The residue numbering for RORα, RORβ, and RORγ are E305-G556, E222-K470, and E269-K518, respectively. Residues around the ligand are shown as red letters. Residues important for ligand binding were labeled on top of the sequences.
Structural model of ROR agonism and antagonism. (A) RORγ agonists, such as 25-hydroxycholesterol, drive recruitment of transcriptional coactivators, which leads to the modulation and promotion of target gene transcription. Inverse agonists of RORγ, such as digoxin, disrupt recruitment of the transcriptional coactivator and repress target gene expression. (B) Agonist binding induces a conformational change and facilitates binding of the LXXLL motif of coactivators, such as SRC2. Antagonists, such as digoxin, induce a conformational change of helix 12 and circumvent the coactivator recruitment. The coactivator protein and helix 12 are colored in red and green, respectively. The agonist (left, 3L0L.pdb) and inverse agonist (right, 3B0W.pdb) are shown as sticks.
Approximately half of the NRs have well-characterized natural ligands, whereas the remaining receptors are classified as orphan NRs because they do not have well-characterized ligands7. Orphan NRs are an active area of research partly due to the potential for clinical agent development for various diseases8. Recent studies have demonstrated that retinoic acid receptor-related orphan receptors (RORs) have been implicated in several physiological and pathological processes. Therefore, RORs have emerged as important drug targets for the treatment of various diseases, such as multiple sclerosis, rheumatoid arthritis, and psoriasis.
Here, we review the structural basis of the ligand regulation mechanism and related diseases, and the strategies to identify potent and specific ROR modulators. The current status of ROR ligand development from both the literature and patents are also described with their therapeutic potentials.
RORs and ROR-related diseases
The ROR subfamily of transcription factors consists of RORα (NR1F1), RORβ (NR1F2) and RORγ (NR1F3) and has been identified in several mammalian species that exhibit tissue-specific expression of these transcription factors9,10. Each ROR gene generates several receptor isoforms that differ in their amino terminus in humans and rodents because of alternative promoter usage and splicing11. The first member of the ROR subfamily of NRs (RORα) was identified in the 1990s based on sequence similarities to the retinoic acid receptor (RAR) and the retinoid X receptor (RXR), which yielded the name 'retinoic acid receptor-related orphan receptor alpha'12. RORβ and RORγ were subsequently identified13,14. RORα, RORβ, and RORγ display distinct patterns of tissue expression. RORα is widely expressed in liver, skeletal muscle, skin, lung, adipose tissue, kidney, thymus, and brain15,16. RORβ exhibits a more restricted neuronal-specific expression pattern in the brain, retina, and pineal gland17,18. RORγ is highly expressed in thymus (the thymus-specific isoform is referred to as RORγt), muscle, testis, pancreas, prostate, heart, and liver10,19. The RORs are somewhat unusual in that they recognize and bind as monomers to specific DNA sequences (typically consisting of TAAA/TNTAGGTCA), termed ROR response elements (ROREs), as opposed to the majority of other NRs, which bind as dimers4,20. When bound to this response element within the promoter of a target gene, RORs constitutively recruit coactivators, which lead to the transcriptional activation of their target genes. By contrast, another group of orphan NRs, the REV-ERBs, repress transcription by recognizing the same response elements and functionally antagonize the action of the RORs in many cases21,22,23. Recent advances have established that selective inhibition of RORs is a promising therapeutic approach for the treatment of autoimmune diseases, metabolic disorders and some cancers24,25,26,27,28,29,30.
RORs and autoimmune diseases
RORα and RORγt (an isoform of RORγ) are considered to be the master regulators of the development of T helper 17 cells (Th17 cells), which have an essential role in the development of many autoimmune disorders, including multiple sclerosis, rheumatoid arthritis, inflammatory bowel disease and psoriasis31,32. Both RORα and RORγt are required for the full differentiation of naïve CD4+ T cells into interleukin 17 (IL-17)-producing Th17 cells33,34,35,36. Th17 cells produce numerous cytokines, including interleukin-17 (IL-17), that are known to enhance inflammatory processes. IL-17A expression is directly regulated by RORs through their interaction with ROREs in the IL-17 promoter33. The discovery of Th17 cells as critical mediators of autoimmune disorders provides a unique opportunity to develop focused therapeutics that act by inhibiting the function of these cells. The genetic ablation of RORγ alone, or in combination with RORα in mice, led to impaired Th17 cell differentiation and protected the mice from the development of experimental autoimmune encephalomyelitis (EAE), a mouse model of multiple sclerosis34. These data suggest that the targeted inhibition of RORα and RORγ with specific synthetic ligands could potentially provide a means for reducing autoimmune pathology. Both RORα and RORγ, specifically RORγt, have gained significant attention over the past few years because of their essential role in the development of Th17 cells. Therefore, pharmacological repression of RORγ might represent an attractive starting point for the development of a novel therapy for treating inflammatory diseases. The current treatments for known Th17-mediated autoimmune diseases use immunosuppressants with significant side effects. Targeting RORs represents a significant advantage over the current therapies because they specifically target the one arm of the immune system that mediates disease instead of the immune system as a whole.
RORs and metabolic disorder
Both RORα and RORγ play important roles in glucose and lipid metabolism, which is exemplified by the phenotypes of mice with mutations in RORα or RORγ37,38. The staggerer mouse (RORαsg/sg) is a natural mutant strain of mouse lacking functional RORα; these animals have lower total plasma cholesterol levels compared with wild-type mice39. Loss of RORα in staggerer mice results in mice that are resistant to weight gain and hepatic steatosis when placed on a high-fat diet38. Suppression of RORα activity may also lead to a decrease in the elevated hepatic glucose output; therefore, RORα inverse agonists may hold utility in the treatment of metabolic disorders, such as type 2 diabetes40,41.
RORγ−/− mice display normal cholesterol and triglyceride levels but slightly reduced blood glucose levels compared with their wild-type counterparts37. In double knockout mice, a similar reduction in cholesterol, triglyceride, and blood glucose levels was observed compared with a single knockout. These findings suggest that RORα and RORγ inverse agonists may hold therapeutic potential for the treatment of metabolic syndrome and associated diseases.
Beyond autoimmunity and metabolic diseases, the RORs also offer the potential for the development of drugs that target a range of disorders, such as asthma and cancer42,43,44.
Structural basis of RORs
A typical NR LBD exhibits similar structural features with a three-layered fold of approximately 12 alpha-helices and 2–3 β-strands. A hydrophobic ligand binding pocket resides within the bottom portion of the LBD (Figure 2B). The helix 12 (also called AF-2) can adopt multiple conformations depending on the different bound ligands (agonist, inverse agonist or antagonist). Therefore, the LBD can interact with a coactivator or a corepressor to activate or repress gene transcription in the nucleus. Upon the binding of an agonist, the helix 12 along with another region of the LBD forms a hydrophobic groove for the binding of a coactivator (such as steroid receptor coactivator, SRC, Figure 2B). The interaction of the LBD with the coactivator will change upon the binding of an inverse agonist (if the NR has constitutive transcriptional activity). Specifically, helix 12 will change its conformation and position relative to the LBD core and therefore no longer interact with either a coactivator or a corepressor (Figure 2B).
The first co-crystal structure of the ROR subfamily was that of RORβ bound with stearic acid (Table 1, 1), a fortuitous ligand45. All-trans retinoic acid (ATRA, 2) and the synthetic retinoid ALTA 1550 (3) were subsequently identified as putative functional ligands and co-crystallized with RORβ46. These ligands function as partial antagonists and inhibit RORβ transcriptional activity. Cholesterol (4) and cholesterol sulfate (5) were subsequently identified by co-crystallization within the ligand binding pocket of RORα in the agonist-bound state, as suggested by a bound coactivator peptide47,48. The first batch of crystal structures of RORγ bound to its agonists 20α-hydroxycholesterol, 22R-hydroxycholesterol and 25-hydroxycholesterol (6, 7, 8) were determined by Jin et al in 201049. These structures revealed that these ligands bind to RORγ in an active conformation with helix 12 positioned for coactivator (SRC2-2) recruitment. In 2011, digoxin (10) was identified as an inverse agonist and the crystal structure of the RORγ LBD in complex with digoxin was determined by Fujita-Sato et al50. To date, at least 3 more complex structures have been solved for the synthetic RORγ agonist (30) and inverse agonists (11, 38). This structural information illustrated the mechanism of action for agonists or inverse agonists. It is very promising that the structure-guided optimization of ligands can facilitate the development of highly selective and potent ROR modulators.
The crystal structures of RORγ LBD bound with compounds 6, 7, and 8 demonstrated that these types of agonists occupy the entire hydrophobic pocket (Figure 3A). They are oriented with the hydroxyl tail toward helix 11 and the A ring toward helixes 1 and 2. The 3β-hydroxyl group of 8 forms a direct hydrogen bond interaction with Gln286 and two water-mediated hydrogen bonds with residues Arg367 and Arg364. The 25-hydroxyl group made one direct hydrogen bond with His479 (helix 11) and one water-mediated hydrogen bond with Try502 (helix 12), thereby stabilizing the π-π interaction network composed of His479, Tyr502 and Phe506 (helix 12). This π-π interaction network, along with Gln487 (helix 11) and Ser507 (helix 12), favors coactivator (SRC2 in this structure) recruitment.
Comparison of the binding of 25-hydroxycholesterol (A), digoxin (B), T0901317 (C), and GSK-2 (D) in the RORγ ligand-binding pocket. Hydrogen bonds are depicted as dashed lines (red). (A) 25-OHC (green) formed direct hydrogen bonds with Gln286 and His479, and water-mediated hydrogen bonds with Arg364, Arg367, and Tyr502. (B) Digoxin (cyan) formed direct hydrogen bonds with Arg367, Phe377, His479, and Leu391, and water-mediated hydrogen bonds with Val361 and Glu379. (C) The phenylsulfonamide group of T0901317 (purple) forms a π−π stacking interaction with Phe378 and Phe388. The ligand and His479 disrupt the interaction network originally formed with Trp317, Tyr502, and Phe506. (D) GSK-2 (orange) formed direct hydrogen bonds with Leu287, Arg367, and Phe377. The amide carbonyl of GSK-2 formed a water-mediated hydrogen bond with Gln286 and His323. The aniline ring of GSK-2 also formed a π−π stacking interaction with Phe378 and Phe388.
The RORγ LBD crystal structure with bound digoxin demonstrated that digoxin forms extensive hydrophobic and hydrophilic interactions with RORγ (Figure 3B). Digoxin forms 4 direct hydrogen bonds with Arg367, Phe377, His479 (helix 11) and Leu391 (helix 6) and 4 water-mediated hydrogen bonds with Glu379 and Val361 (helix 5). Digoxin disrupts the polar interaction observed in the 8-bound RORγ LBD, involving His479, Tyr502, and Phe506, which could be important to stabilize the active agonist conformation. Digoxin binding also induces a side chain conformational change in Trp317 (helix 3), which could cause the large movement of helix 12. It is clear that digitoxose Y and Z protrudes between helixes 3 and 11, and prevents the positioning of helix 12 in the active conformation, as observed in the RORγ LBD complexes with 8.
The benzenesulfonamide compound T0901317 (11) was initially identified as a potent agonist of the liver X receptor51. When a team from the Scripps Research Institute in Florida performed a screen to profile the activity of a collection of well-characterized NR ligands against all human NRs, they identified T0901317 as a potent inverse agonist of RORα and RORγ. Fauber et al from Genentech, Inc obtained the complex crystal structure of T0901317 and RORγ. This structure provided a detailed molecular insight into why T0901317 functioned as an inverse agonist of RORγ but an agonist of FXR, LXR, and PXR. There are several unique structural features in this complex structure in comparison with the structures of other complexes (Figure 3C). T0901317, because of its small size, does not occupy the polar pocket formed by Try281, Gln286, Arg364, Arg367, and Phe378. The phenylsulfonamide group of T0901317 forms a unique π–π stacking interaction with residues Phe378 and Phe388. This interaction was not observed in the structures of LXR and PXR in complex with T9091317, or RORγ in complex with other hydroxycholesterol and digoxin derivatives. The hexafluoroisopropanol hydroxyl group forms a strong hydrogen bond interaction with the His421 residue in LXRα, which results in stabilization of helixes 11 and 12 and the recruitment of the coactivator peptide. However, there are no obvious hydrogen bonds from this hydroxyl group to the protein. The His479 residue undergoes a conformational change because one of the trifluoromethyl groups occupies its original position. In this case, the ligand and His479 destabilize the interaction network that was originally formed with Trp317, Try502, and Phe506, and disrupt coactivator recruitment. This effect may also explain why T0901317 behaved as an inverse agonist of RORγ.
A team from GlaxoSmithKline developed a series of tertiary amine derivatives as RORγ inverse agonists52,53. However, they only determined the structure of RORγ with agonist 30 (Figure 3D). From this structure, one can clearly determine that this series of ligands form several direct or water-mediated hydrogen bonds. The sulfone moiety forms hydrogen bonds with Arg367 and Leu287. The amide forms one direct hydrogen bond with Phe377 and water-mediated hydrogen bonds with Glu286 and His323. The phenyl group interacts with Trp317 and His479 and therefore stabilizes the network with Try502 and Phe506 in the active conformation. However, the 4-CF3-phenyl group will disrupt this interaction, and helix 12 cannot maintain its active conformation52. This finding may explain why a close derivative with the 4-CF3-phenyl group behaved as an inverse agonist of RORγ, whereas 30 was an agonist. There is another published crystal structure by Genentech (4QM0.pdb)54.
All of these crystal structures of the RORγ LBD in complex with its ligands provide the basis for rational drug design to obtain more potent and specific RORγ modulators.
Strategies for the discovery of NR ligands
The identification of potential drug candidates for NRs represents a promising therapeutic approach to various diseases. Several drug development strategies have been developed to identify compounds that bind to NR LBD. Some approaches are rapid, accurate and easy to be developed in a high-throughput screen format to measure the NR-ligand interaction. These strategies are described below.
AlphaScreen assay
AlphaScreen, a bead-based Amplified Luminescent Proximity Homogeneous Assay, was first described in 1994 by Ullman based on the principle of luminescent oxygen channeling55,56. The donor and acceptor beads are brought into close proximity when a molecular interaction of binding partners immobilized to these beads occurs (Figure 4A). The excitation of the assay mixture with a high intensity laser at 680 nm initiates a luminescence/fluorescence cascade in the acceptor beads and leads to a highly amplified signal with light output at 520–620 nm. When the acceptor and donor beads are not in proximity, only a very low background signal is generated. The major advantages of AlphaScreen include the high sensitivity, large signal/background ratio, and low reaction volumes (25–40 μL). AlphaScreen has become one of the best methods for the high-throughput screening for the detection of receptor-ligand interactions, such as ligand-induced NR-coregulator interactions57. Another advantage of AlphaScreen is that it can distinguish between an agonist and an antagonist using different coactivator or corepressor peptides.
Representative drug discovery strategies. (A) Schematic representation of the AlphaScreen assay (Amplified Luminescent Proximity Homogenous Screen Assay Screen). H6-RORγ is immobilized on Ni-chelating acceptor beads and the biotinylated coactivator (Biotin-SRC) on streptavidin-coated donor beads. Donor beads contain a photosensitizer that, upon activation at 680 nm, converts ambient oxygen to singlet oxygen. If the acceptor beads are brought into close proximity of the donor beads by a RORγ-coactivator interaction, energy is transferred from the singlet oxygen to the thioxene derivatives in the acceptor beads, which results in light emission at 520–620 nm. Addition of a RORγ inverse agonist represses the signal of acceptor bead-immobilized H6-RORγ and donor-bead-biotin-coactivator. (B) Schematic representation of fluorescence intensity versus temperature of the melting protein in the presence of SYPRO orange. Ligand binding to a target protein can stabilize a protein's native state reflected in the increased melting temperature (Tm) of the bound protein. Monitoring of the ΔTm of apo and ligand-bound proteins can be used to determine the ligand binding affinity. (C) Cell-based reporter assays. [left] Ligand binds to the NR LBD and the NR DBD binds to the nuclear receptor response element upstream of the reporter gene to activate transcription. [right] Upon ligand binding, the GAL4-NR-LBD binds to the GAL4 UAS to activate transcription.
An increasingly greater number of natural or synthesized NR ligands have been identified by AlphaScreen49,58,59,60. Using Ni-chelating acceptor beads and streptavidin-coated donor bead pairs from PerkinElmer, a his-tag fusion NR protein and biotinylated coregulator peptides, we also found some NR modulators. These ligands include a PPARγ agonist61, an orphan receptor RORγ antagonist62, and natural compounds as agonists for the orphan receptors TR4 and COUP-TFII63,64.
DSF
Differential scanning fluorimetry (DSF), also known as the protein thermal shift assay (TSA), is an increasingly popular method to identify specific ligands or nonspecific protein stabilizing conditions, such as buffers, salts, pH and small molecule ligands65,66,67. TSA monitors the thermal unfolding of proteins in the presence of an environmentally sensitive fluorescent dye, such as SYPRO Orange, and measures the melting temperature (Tm) as a readout (Figure 4B). The SYPRO Orange dye is highly fluorescent in a non-polar environment (hydrophobic core residues exposed to the dye), whereas the fluorescence is quenched in aqueous surroundings. The melting temperature is the midpoint of transition from native to unfolded state and provides information on the thermal stability of a protein in apo or holo form.
DSF is a rapid and inexpensive screening method to identify ligands that bind and stabilize proteins. It has been demonstrated that the stabilizing effect of a ligand upon binding is proportional to the concentration and affinity of the ligand68,69,70. Compounds with similar physicochemical properties can be ranked based on their relative ΔTm values71. DSF is compatible with a standard real-time PCR instrument and can be performed in 96-well format using 10 μL reaction volumes and with a small amount of protein required. DSF has received substantial attention and has been widely applied for the development of drug candidates in recent years70,72,73.
DeSantis and Sekiguchi et al successfully identified ERα and PXR ligands using DSF as a high-throughput assay74,75. Our group developed a high-throughput assay for RORγ ligand identification using DSF. The positive control digoxin exhibited a 10 °C temperature shift compared with DMSO (Tm=66 °C for digoxin and Tm=56 °C for DMSO). A series of benzenesulfonamide-containing compounds (19) were identified from DSF screening of our in-house chemical library and could stabilize RORγ by 5–7 °C (unpublished data).
Cell-based reporter assay
A cell-based reporter assay may provide valuable information regarding the functional activity, potency and selectivity of a ligand that interacts with NRs. Transient and stable transfections are two types of cell-based systems for evaluating NR transactivation, and the former is the most common. The NR LBD fused with a GAL4-DBD is cotransfected with a reporter construct that contains GAL4 upstream activation sequences (UAS) of a reporter gene such as luciferase (Figure 4C). The regulation of transactivation of the NR by binding of a ligand may be measured by the expression level of the reporter gene. For native promoter reporter assays, the cells are cotransfected with plasmids that encode a full-length NR and their cognate luciferase reporters such as RORE-luc (Figure 4C). There are many modified versions of gene reporter assay systems for the measurement of receptor activation or repression. These systems are easily used to rapidly determine compound selectivity and potency in NRs76,77,78,79. Similar to the AlphaScreen assay, the cell-based transactivation assay can distinguish between an agonist and an antagonist.
Virtual screening approaches
Virtual screening (VS) is a knowledge-based approach which can be divided into structure-based and ligand-based methods. For structure-based virtual screening, the most frequently used method is molecular docking when the 3D structure of the target is available. The ligand-based methods have also been widely used because it is unnecessary to know 3D information for the targets. There have been several well-established methods demonstrated in the literature, such as 3D-QSAR, ligand-based pharmacophore searching, scaffold-hopping, virtual library design, and shape-based screening. Readers are encouraged to refer to the excellent reviews by Jiang et al for various computational drug design methods80,81. Virtual screening has been widely used for the discovery of NR ligands62,82,83,84,85.
Beyond the strategies previously described, fluorescence resonance energy transfer (FRET) or time-resolved FRET has also been used as a high-throughput tool for the discovery of NR modulators52,86. Furthermore, differential hydrogen/deuterium exchange mass spectrometry (H/D-Ex) has been used to monitor the conformational change from the apo-form to ligand-bound form of a NR LBD29,41.
Discovery of ROR modulators
The RORs were initially identified as orphan receptors, and it was not clear that these receptors were regulated by small molecule ligands. The co-crystal structure of the LBD of RORα bound to cholesterol indicated that various cholesterol derivatives, such as 7-oxygenated sterols, may act as physiological ligands to influence ROR activity. Given the specific tissue distribution of each ROR isoform and their potential role in pathophysiological conditions, they have been linked to autoimmune diseases and metabolic disorders, as previously described. The discovery and development of natural and synthetic ligands, including agonists, antagonists, and inverse agonists that modulate the activity of these receptors is in high demand.
Ursolic acid (UA, 9), a natural carboxylic acid ubiquitously present in plants, was identified as a strong and selective inhibitor for RORγt function86. UA was identified from a small chemical library with more than 2000 known bioactive compounds. UA inhibited IL-17 production not only in developing Th17 cells but also in mature Th17 cells. Mice that received UA treatment were resistant to EAE, which indicates UA can be used to develop a treatment for Th17-mediated diseases. UA was demonstrated to inhibit the binding of RORγ-LBD to coactivator peptide SRC-1 with an IC50 of 0.68 μmol/L. Consistent with the results of retroviral overexpression and reporter gene assays, UA did not inhibit the binding of RORα-LBD to the coactivator peptide, which suggests that UA is a RORγt-specific antagonist. As a natural triterpene scaffold molecule, ubiquitously present in plants and human diets, UA is relatively non-toxic and is well tolerated both orally and topically in humans and rodents. These characteristics suggest that UA has a great advantage in the development of more selective RORγ modulators.
Digoxin (10), a well-known cardiac glycoside used clinically in the treatment for various heart conditions, was identified as a specific inhibitor for RORγ in 201130,50. Huh et al performed a chemical screen with 4812 compounds and determined that digoxin inhibited RORγ transcriptional activity in a Drosophila S2 cell luciferase reporter assay with an IC50 value of 1.98 μmol/L. Digoxin inhibition of RORγ was specific, with no activity against RORα, DAF-12, AR, or LXRα. Digoxin can directly bind to RORγ-LBD and displace 25-hydroxycholesterol (8) with an IC50 of 4.1 μmol/L. Circular dichroism analysis also demonstrated that digoxin increased the thermal stability of RORγ-LBD. Digoxin inhibited murine Th17 cell differentiation without affecting the differentiation of other T cell lineages, including Th1, Th2, and regulatory T cells. Digoxin was effective in delaying the onset and reducing the severity of symptoms in a mouse model of multiple sclerosis (EAE). Digoxin is unlikely to be used therapeutically for inflammatory and autoimmune diseases because of its toxicity. However, it may still serve as a good template for the development of a safer RORγ antagonist.
In 2010, the Griffin and Burris laboratories at Scripps Florida demonstrated that the benzenesulfonamide LXR agonist T0901317 also functions as a potent inverse agonist of RORα and RORγ via suppression of their basal transcriptional activity87. Griffin et al demonstrated, for the first time, that a synthetic ligand could bind directly to and modulate the transcriptional activity of RORα and RORγ with high affinity (Ki=132 and 51 nmol/L for RORα and RORγ, respectively). It is interesting that T0901317 inhibited the constitutive transactivation activity of both GAL4-RORα and GAL4-RORγ with little or no activity on GAL4-RORβ. In control cells transfected with GAL4-VP16 and the UAS reporter, no repression of GAL4-VP16 transactivation of the luciferase gene was observed, which suggests that the repression induced by T0901317 is not a result of nonspecific luciferase effects or cellular toxicity. Treatment of cells that expressed GAL4-LXRα, GAL4-RORα, or GAL4-RORγ with increasing concentrations of T0901317 demonstrated an excellent dose response, with an estimated EC50 of 0.25 μmol/L (LXRα) and estimated IC50 values of 2.0 μmol/L (RORα) and 1.7 μmol/L (RORγ). Medicinal chemistry efforts focused on the T0901317 scaffold led to the development of a series of non-sterol, synthetic ROR modulators.
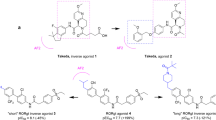
In a 2011 patent, Scripps Research Institute worked on the T0901317 scaffold to explore SAR in search of selective RORγ ligands. They determined that SR1001 (12) targets both RORα and RORγ and that both receptors are required for the development of Th17 cell-mediated autoimmune diseases29,33,34. SR1078 (13) was the first identified agonist of RORα and RORγ88. SR3335 (14) was the first potent RORα-specific inverse agonist41. Additional work on the SR1001 scaffold, directed at the design of RORγ-selective inverse agonists, led to the identification of two RORγ-specific ligands, SR1555 (15) and SR2211 (16)89,90.
SR1001 (12) has a hexafluoroisopropanol-substituted phenyl group with an N-linked sulfonamide in the para position (Figure 5A). There are some major structural differences between SR1001 and T0901317. A more elaborate substituted thiazole is attached to the sulfonamide sulfur in SR1001; however, a phenyl ring is attached to this portion in T0901317. The trifluoroethyl group on the sulfonamide nitrogen in T0901317 was simplified to an N-H in SR1001. SR1001 repressed both GAL4–RORα and GAL4–RORγ transcriptional activities in a dose-dependent manner, but demonstrated no effect on LXRα activity. The specificity of SR1001 was also assessed in a panel that comprised all 48 human NRs in a cell-based cotransfection assay, and no activity was observed on receptors other than RORα or RORγ. The direct binding of SR1001 to RORα and RORγ was examined using competitive radioligand binding assays. SR1001 dose-dependently displaced [3H]25-hydroxycholesterol binding to RORα and RORγ (Ki=172 and 111 nmol/L, respectively). SR1001 reduced the interaction of a co-activator TRAP220 NR box 2 peptide with RORγ in a dose-dependent manner (IC50=117 nmol/L).
Structures of RORγ inverse agonists from scripps institute (A), GSK (B), and Japan tobacco (C).
SR1078 (13) contained the same hexafluoroisopropanol carboxylic acid isostere as T0901317 but used a secondary amide, rather than a tertiary sulfonamide, to link the two phenyl rings. In an AlphaScreen assay, increasing doses of SR1078 resulted in a dose-dependent reduction in the ability of RORγ to recruit the TRAP220 coactivator NR box. In a cell-based GAL4-NRLBD cotransfection assay, SR1078 significantly inhibited the constitutive transactivation activity of RORα and RORγ but had no effect on the activity of FXR, LXRα, or LXRβ. These data clearly demonstrate that SR1078 selectively targeted RORα and RORγ and no longer functioned as a LXR/FXR agonist.
Additional modifications of the T0901317 and SR1078 scaffolds led to the discovery of SR3335 (14), the first potent RORα-specific inverse agonist. SR3335 was initially identified by its ability to inhibit the constitutive activity of RORα in a GAL4-RORα-LBD cotransfection assay. In a biochemical radioligand binding assay that used [3H]25-hydroxycholesterol as a label, it was clear that unlabeled SR3335 dose-dependently competed for binding to the RORα LBD (Ki=220 nmol/L). SR3335 did not compete well for binding when the RORγ LBD was utilized. In a cell-based cotransfection assay, SR3335 significantly inhibited the constitutive transactivation activity of RORα (IC50=480 nmol/L) (partial inverse agonist activity) but had no effect on the activity of LXRα or RORγ.
Using a modular chemistry approach, modifications to the SR1001 scaffold were performed to develop SAR to diminish RORα activity while maintaining selectivity over LXR. The compounds were profiled using a screening approach based on radioligand binding in a Scintillation Proximity Assay (SPA) format. SR2211 (16) displays exquisite selectivity for RORγ over RORα in both biochemical and cell-based assays, with a Ki value of 105 nmol/L at RORγ and no detectable binding to RORα. To assess the functional transcriptional activity of SR2211, cell-based assays using GAL4-NR-LBD cotransfection assays (LBDs of RORα, RORγ, LXRα, FXR, and VP-16) were performed. SR2211 treatment did not have an impact on the transcriptional activity of RORα, whereas the inhibition of RORγ activity was observed with an IC50 of 320 nmol/L.
A screen of SR1001 derivatives in a GAL4-NR co-transfection assay demonstrated that SR1555 (15) was devoid of LXR, FXR, and RORα activity but that it repressed the activity at RORγ in a dose-dependent manner (IC50≈1.5 μmol/L). SR1555 was only able to displace [3H]T0901317 from the ligand binding domain (LBD) of RORγ (IC50=1.0 μmol/L) and not RORα, which confirmed that SR1555 was indeed specific for RORγ.
To identify selective RORγ antagonists, Scripps Research Institute performed a quantitative high-throughput screen of 310 000 compounds using a cell-based RORγ gene reporter assay to detect RORγ transcriptional inhibitors. The initial hit displayed an IC50 value of 3.3 μmol/L in the RORγ assay with no activity in control assays, including the RORα assay. The initial hit also inhibited Th17 cell differentiation, which indicated that this was a viable scaffold for the development of SAR studies. These efforts led to the discovery of ML209 (SR9805, 17), which is a RORγ inhibitor with an IC50 value of 0.5 μmol/L91,92. Notably, in a panel of 20 NRs, only weak activity was reported for ERRα, LXRα, and thyroid hormone receptor α and β (TRα and TRβ). In accordance with the cell-based RORγ gene reporter assay, ML209 was 12-fold more potent (IC50=51 nmol/L) compared with its enantiomer (IC50=605 nmol/L) in the competition assays91,92.
In 2012, a team from Innovimmune disclosed a series of related indoline and tetrahydroquinoline modulators of RORγ. The SAR for these compounds appeared to be tight, with only subtle structural variations giving rise to RORγ agonist or inverse agonist activity, as assessed by a coactivator peptide recruitment assay. Compound 18 described in these patents bears some resemblance to T0901317. All compounds displayed a modest inhibition of IL-17A and IL-17F production in a human PBMC assay (EC50<10 μmol/L). The compounds were identified as potent RORγ inverse agonists in a TR-FRET assay (EC50<30 μmol/L). It will be interesting to determine whether this recently disclosed set of compounds from Innovimmune are selective for RORγ over LXR93.
N-Acylated tetrahydroquinolines, indazoles, benzoxazine arylsulfonamides and related analogues have recently been reported in patents as potential inverse agonists of RORγ. Nonetheless, there are several noticeable trends throughout these series. First, the N-1 amine of the tetrahydroquinoline, indazole, and benzoxazine rings is preferably sulfonylated. It is not clear what the nature of this effect is, other than to force the rings out of co-planarity, but this type of substitution dominates the majority of the examples. Compound 20 from Lycera's patent contained a 2,6-disubstituted benzamide at the 7-position of the tetrahydroquinoline core94. These compounds are claimed as RORγ inhibitors for reducing IL-17 and treating immune and inflammatory disorders. The compounds were tested for their ability to inhibit RORγ activity in cells using a GAL4-driven reporter gene assay in HEK293 cells. The compounds were also tested for binding in a RORγ-LBD TR-FRET assay. No clear data are provided other than whether the IC50 values are less than or greater than 15 μmol/L, which made it impossible to decipher any SAR. To further explore SAR, compound 21 was found to exhibit an EC50 of 6 nmol/L in a SRC1-2 coactivator peptide recruitment assay95.
Merck disclosed a series of indazoles and azaindazoles as inverse agonists of RORγ. A series of sulfonylated indazoles were described in a European patent application from Merck. Two of the most potent compounds, 22 and 2396, exhibited IC50 values of 2 nmol/L. This series is similar to that from Lycera, in which the 2,6-bishalogenated benzoyl substitution pattern appears on the indazole nitrogen atom in this case. Although the claims indicate the core can be an indole, all examples described are indazoles. This series also appears to tolerate sulfonyl substitution on the indazole nitrogen, as several potent analogs of this type are also described. Indazole ring substitution, as well as aza-analogs, is tolerated.
Our group also identified N-acylated tetrahydroquinolines, indazoles, benzoxazine arylsulfonamides and related analogues in the TSA screening. Compound 19 was demonstrated to have an IC50 value of less than 1.0 μmol/L using a GAL4-driven reporter gene assay in HEK293 cells (unpublished data).
GSK has conducted substantial research on the discovery of RORγ modulators. A series of aromatic amide derivatives were identified as agonists of RORγ using a TR-FRET assay. Compounds 24, 25, and 26 were evaluated for SRC1 recruitment. These compounds were also demonstrated to stimulate RORγ-dependent IL-17 expression in a luciferase reporter assay with an EC50 value of ∼100 nmol/L. These compounds were demonstrated to directly interact with RORγ using a thermal stability shift assay measured by circular dichroism97. Compounds 24, 25, and 26 stabilized RORγ by 0.49, 2.26, and 2.90 °C, respectively.
In 2012, high-throughput screening of the GSK in-house compound collection using a FRET assay resulted in the identification of thiazole amide compound 27 as a RORγ inhibitor with a pIC50 of 6.0. The binding of compound 27 to the RORγt LBD was confirmed with a thermal shift of 7.1 °C in a thermal shift assay and a pKi of 6.4 in a radioligand binding assay. In the subsequent evaluation in a cell-based assay, compound 27 inhibited Th17 cell differentiation with 49% of maximum inhibition at 10 μmol/L. In all patent applications, the 2-(4-(ethylsulfonyl)phenyl)acetamide group of 27 was fixed, and the SAR study was explored on the right-hand side with the thiazole amide as a core structure. This moiety appears to be critical for activity. The central core of compound 27 is always thiazole, or thiophene. SAR studies of compound 27 led to the identification of thiazole ketone amide compound 28 and thiophene ketone amide compound 29 with high binding affinities and inhibitory activities for Th17 cell differentiation. For example, compound 28 exhibited a FRET pIC50 of 7.8 and a Th17 pIC50 of 6.7. Compound 29 exhibited excellent inhibitory activity on a Th17 cell differentiation assay with a pIC50 of 7.9, and in a RORγ FRET assay with a pIC50 of 7.8. The reason why thiazole ketone amides exhibited lower Th17 potency compared with RORγ FRET potency is unclear and cannot be explained by membrane permeability because both thiazole ketone amides and thiophene ketone amides have reasonably good membrane permeability53.
SAR exploration on the right-hand side (RHS) of compound 27 (Figure 5B) led to the identification of tertiary amine compound 30 as a potent RORγt agonist with an EC50 of 20 nmol/L in a dual FRET assay. Following this assessment, compound 31 was synthesized and confirmed to be a potent RORγt inverse agonist with an IC50 of 5 nmol/L (FRET, max%=104). Linker exploration and rational design led to a series of indole-based analogues as more potent RORγt inverse agonists. Compound 32 was discovered as a potent RORγt lead with an IC50 less than 10 nmol/L in a FRET assay53.
In a separate but somewhat related patent, Glenmark disclosed a series of biaryl and heterobiaryl amide analogues as modulators of RORγ. In patent applications, the 4-substituted ethyl sulfone phenacetyl amide is constant. These compounds were strikingly similar to the compounds described in 2013 by the team at GSK. Compounds 33 and 34 from Glenmark were described and tested in a TR-FRET assay and found to have IC50 values <50 nmol/L98.
Another patent disclosure from GSK described a series of N-aryl benzenesulfonamides as RORγ inverse agonists. Compounds 35, 36, and 3799,100 from the patent were potent RORγ inverse agonists with pIC50 values >7.8 in the SRC1-2 coactivator peptide recruitment assay. These compounds also inhibited IL-17 production in a human PBMC assay (pIC50>6.0). Two follow-up patents on the same series were disclosed and exemplified subtle changes on the periphery of the compounds. Compound 35 was reported as a racemate, and its separate enantiomers were also described in the same patent. Compound 35 (the racemate) was a potent inhibitor with a pIC50=7.7 in a TR-FRET assay and a pIC50=7.5 in a human PBMC assay.
Genentech recently described a series of sulfonamides as modulators of RORγ. The compounds were assessed for their ability to bind to the RORγ LDB by displacing [3H]25-hydroxycholesterol in a radioligand binding assay. These compounds were strikingly similar compared with the compounds described earlier by the team at GSK. Compound 38 was demonstrated to be a potent inhibitor with an IC50 of 130 nmol/L. Compound 39 demonstrated inhibition activity of RORγ LBD recruitment of a peptide derived from the SRC1 co-activator protein (EC50=57 nmol/L), which was shown in GAL4 cellular constructs to have an EC50 value of 120 nmol/L54,101.
Our group recently discovered a new series of sulfonamide RORγ antagonists using a structure-based virtual screening approach in conjunction with medicinal chemistry optimization and biological evaluation. The derivatives were synthesized or purchased, and assessed with the AlphaScreen assay and luciferase reporter gene assays. The derivatives demonstrated remarkably improved activity. The most potent was compound 40, which showed an IC50 value of 50 nmol/L in a GAL4-driven luciferase reporter assay62.
One patent from Japan Tobacco disclosed a series of isoxazole- or triazole-based compounds as RORγ inverse agonists. Some of the reported compounds in the patent exhibited EC50 values less than 3.0 μmol/L in a GAL4-driven luciferase reporter assay102. Japan Tobacco recently announced a phase I clinical trial of a RORγ inverse agonist, JTE-151 (Figure 5C). This trial will provide a detailed description of the long-term in vivo safety profiles of this RORγ inverse agonist.
Conclusions
Orphan NRs are potential drug targets, and their functions can be regulated by potent and specific agonists or inverse agonists. The identification of the natural ligands for the retinoic acid receptor-related orphan receptors remains controversial, although a range of oxysterols can potently bind to and modulate the function of RORγ. Since the benzenesulfonamide LXR agonist T0901317 was identified as an inverse agonist of both RORα and RORγ, significant progress have been made regarding the identification of novel ligands for the RORs. It is obvious that the determination of more complex structures of RORs and their ligands can facilitate the structure-based design of potent and selective ROR modulators. Further optimization for improved drug-like properties of these compounds will have a high potential for the treatment of metabolic and autoimmune diseases.
References
Mangelsdorf DJ, Evans RM . The RXR heterodimers and orphan receptors. Cell 1995; 83: 841–50.
Evans RM . The steroid and thyroid hormone receptor superfamily. Science 1988; 240: 889–95.
Burris TP, Solt LA, Wang Y, Crumbley C, Banerjee S, Griffett K, et al. Nuclear receptors and their selective pharmacologic modulators. Pharmacol Rev 2013; 65: 710–78.
Giguere V . Orphan nuclear receptors: from gene to function. Endocr Rev 1999; 20: 689–725.
Laudet V . Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J Mol Endocrinol 1997; 19: 207–26.
Overington JP, Al-Lazikani B, Hopkins AL . How many drug targets are there? Nat Rev Drug Discov 2006; 5: 993–6.
Hummasti S, Tontonoz P . Adopting new orphans into the family of metabolic regulators. Mol Endocrinol 2008; 22: 1743–53.
Mohan R, Heyman RA . Orphan nuclear receptor modulators. Curr Top Med Chem 2003; 3: 1637–47.
Benoit G, Cooney A, Giguere V, Ingraham H, Lazar M, Muscat G, et al. International union of pharmacology. LXVI. Orphan nuclear receptors. Pharmacol Rev 2006; 58: 798–836.
Jetten AM . Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal 2009; 7: e003.
Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science 2000; 288: 2369–73.
Giguere V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G . Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev 1994; 8: 538–53.
Carlberg C, Hooft van Huijsduijnen R, Staple JK, DeLamarter JF, Becker-Andre M . RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Mol Endocrinol 1994; 8: 757–70.
Hirose T, Smith RJ, Jetten AM . ROR gamma: the third member of ROR/RZR orphan receptor subfamily that is highly expressed in skeletal muscle. Biochem Biophys Res Commun 1994; 205: 1976–83.
Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, et al. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature 1996; 379: 736–9.
Steinmayr M, Andre E, Conquet F, Rondi-Reig L, Delhaye-Bouchaud N, Auclair N, et al. staggerer phenotype in retinoid-related orphan receptor alpha-deficient mice. Proc Natl Acad Sci U S A 1998; 95: 3960–5.
Andre E, Gawlas K, Becker-Andre M . A novel isoform of the orphan nuclear receptor RORbeta is specifically expressed in pineal gland and retina. Gene 1998; 216: 277–83.
Andre E, Conquet F, Steinmayr M, Stratton SC, Porciatti V, Becker-Andre M . Disruption of retinoid-related orphan receptor beta changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J 1998; 17: 3867–77.
Medvedev A, Yan ZH, Hirose T, Giguere V, Jetten AM . Cloning of a cDNA encoding the murine orphan receptor RZR/ROR gamma and characterization of its response element. Gene 1996; 181: 199–206.
Jetten AM, Kurebayashi S, Ueda E . The ROR nuclear orphan receptor subfamily: critical regulators of multiple biological processes. Prog Nucleic Acid Res Mol Biol 2001; 69: 205–47.
Burris TP . Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol Endocrinol 2008; 22: 1509–20.
Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol 2007; 14: 1207–13.
Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 2007; 318: 1786–9.
Chang MR, Lyda B, Kamenecka TM, Griffin PR . Pharmacologic repression of retinoic acid receptor-related orphan nuclear receptor gamma is therapeutic in the collagen-induced arthritis experimental model. Arthritis Rheumatol 2014; 66: 579–88.
Burris TP, Busby SA, Griffin PR . Targeting orphan nuclear receptors for treatment of metabolic diseases and autoimmunity. Chem Biol 2012; 19: 51–9.
Chang MR, Rosen H, Griffin PR . RORs in autoimmune disease. Curr Top Microbiol Immunol 2014; 378: 171–82.
Marciano DP, Chang MR, Corzo CA, Goswami D, Lam VQ, Pascal BD, et al. The therapeutic potential of nuclear receptor modulators for treatment of metabolic disorders: PPARgamma, RORs, and Rev-erbs. Cell Metab 2014; 19: 193–208.
Solt LA, Burris TP . Action of RORs and their ligands in (patho)physiology. Trends Endocrinol Metab 2012; 23: 619–27.
Solt LA, Kumar N, Nuhant P, Wang Y, Lauer JL, Liu J, et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature 2011; 472: 491–4.
Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature 2011; 472: 486–90.
Korn T, Bettelli E, Oukka M, Kuchroo VK . IL-17 and Th17 cells. Annu Rev Immunol 2009; 27: 485–517.
Tesmer LA, Lundy SK, Sarkar S, Fox DA . Th17 cells in human disease. Immunol Rev 2008; 223: 87–113.
Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity 2008; 28: 29–39.
Ivanov, II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 2006; 126: 1121–33.
Ivanov, II, Zhou L, Littman DR . Transcriptional regulation of Th17 cell differentiation. Semin Immunol 2007; 19: 409–17.
Manel N, Unutmaz D, Littman DR . The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat Immunol 2008; 9: 641–9.
Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, et al. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genomics 2007; 31: 281–94.
Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE . The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem 2008; 283: 18411–21.
Mamontova A, Seguret-Mace S, Esposito B, Chaniale C, Bouly M, Delhaye-Bouchaud N, et al. Severe atherosclerosis and hypoalphalipoproteinemia in the staggerer mouse, a mutant of the nuclear receptor RORalpha. Circulation 1998; 98: 2738–43.
Chopra AR, Louet JF, Saha P, An J, Demayo F, Xu J, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke's disease. Science 2008; 322: 1395–9.
Kumar N, Kojetin DJ, Solt LA, Kumar KG, Nuhant P, Duckett DR, et al. Identification of SR3335 (ML-176): a synthetic ROR alpha selective inverse agonist. ACS Chem Biol 2011; 6: 218–22.
Zhu Y, McAvoy S, Kuhn R, Smith DI . RORA, a large common fragile site gene, is involved in cellular stress response. Oncogene 2006; 25: 2901–8.
Wang Y, Solt LA, Kojetin DJ, Burris TP . Regulation of p53 stability and apoptosis by a ROR agonist. PLoS ONE 2012; 7: e34921.
Du J, Xu R . RORalpha, a potential tumor suppressor and therapeutic target of breast cancer. Int J Mol Sci 2012; 13: 15755–66.
Stehlin C, Wurtz JM, Steinmetz A, Greiner E, Schule R, Moras D, et al. X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation. EMBO J 2001; 20: 5822–31.
Stehlin-Gaon C, Willmann D, Zeyer D, Sanglier S, Van Dorsselaer A, Renaud JP, et al. All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR beta. Nat Struct Biol 2003; 10: 820–5.
Kallen J, Schlaeppi JM, Bitsch F, Delhon I, Fournier B . Crystal structure of the human ROR alpha ligand binding domain in complex with cholesterol sulfate at 2.2 angstrom. J Biol Chem 2004; 279: 14033–38.
Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, et al. X-ray structure of the hROR alpha LBD at 1.63 angstrom: Structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of ROR alpha. Structure 2002; 10: 1697–707.
Jin L, Martynowski D, Zheng S, Wada T, Xie W, Li Y . Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol Endocrinol 2010; 24: 923–9.
Fujita-Sato S, Ito S, Isobe T, Ohyama T, Wakabayashi K, Morishita K, et al. Structural basis of digoxin that antagonizes ROR gamma t receptor activity and suppresses Th17 cell differentiation and interleukin (IL)-17 production. J Biol Chem 2011; 286: 31409–17.
Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, et al. Role of LXRs in control of lipogenesis. Genes Dev 2000; 14: 2831–8.
Yang T, Liu Q, Cheng YB, Cai W, Ma YL, Yang LQ, et al. Discovery of tertiary amine and indole derivatives as potent ROR gamma t inverse agonists. ACS Med Chem Lett 2014; 5: 65–8.
Wang YH, Cai W, Zhang GF, Yang T, Liu Q, Cheng YB, et al. Discovery of novel N-(5-(arylcarbonyl)thiazol-2-yl) amides and N-(5-(arylcarbonyl)thiophen-2-yl)amides as potent ROR gamma t inhibitors. Bioorg Med Chem 2014; 22: 692–702.
Fauber BP, Rene O, de Leon Boenig G, Burton B, Deng Y, Eidenschenk C, et al. Reduction in lipophilicity improved the solubility, plasma-protein binding, and permeability of tertiary sulfonamide RORc inverse agonists. Bioorg Med Chem Lett 2014; 24: 3891–7.
Ullman EF, Kirakossian H, Switchenko AC, Ishkanian J, Ericson M, Wartchow CA, et al. Luminescent oxygen channeling assay (LOCI): sensitive, broadly applicable homogeneous immunoassay method. Clin Chem 1996; 42: 1518–26.
Ullman EF, Kirakossian H, Singh S, Wu ZP, Irvin BR, Pease JS, et al. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence. Proc Natl Acad Sci U S A 1994; 91: 5426–30.
Glickman JF, Wu X, Mercuri R, Illy C, Bowen BR, He Y, et al. A comparison of ALPHAScreen, TR-FRET, and TRF as assay methods for FXR nuclear receptors. J Biomol Screen 2002; 7: 3–10.
Wang S, Wang Z, Lin S, Zheng W, Wang R, Jin S, et al. Revealing a natural marine product as a novel agonist for retinoic acid receptors with a unique binding mode and inhibitory effects on cancer cells. Biochem J 2012; 446: 79–87.
Jin L, Feng X, Rong H, Pan Z, Inaba Y, Qiu L, et al. The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism. Nat Commun 2013; 4: 1937.
Jin L, Lin S, Rong H, Zheng S, Jin S, Wang R, et al. Structural basis for iloprost as a dual peroxisome proliferator-activated receptor alpha/delta agonist. J Biol Chem 2011; 286: 31473–9.
Malapaka RR, Khoo S, Zhang J, Choi JH, Zhou XE, Xu Y, et al. Identification and mechanism of 10-carbon fatty acid as modulating ligand of peroxisome proliferator-activated receptors. J Biol Chem 2012; 287: 183–95.
Zhang Y, Xue X, Jin X, Song Y, Li J, Luo X, et al. Discovery of 2-oxo-1,2-dihydrobenzo[cd]indole-6-sulfonamide derivatives as new RORgamma inhibitors using virtual screening, synthesis and biological evaluation. Eur J Med Chem 2014; 78: 431–41.
Zhou XE, Suino-Powell KM, Xu Y, Chan CW, Tanabe O, Kruse SW, et al. The orphan nuclear receptor TR4 is a vitamin A-activated nuclear receptor. J Biol Chem 2011; 286: 2877–85.
Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, et al. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 2008; 6: e277.
Fedorov O, Niesen FH, Knapp S . Kinase inhibitor selectivity profiling using differential scanning fluorimetry. Methods Mol Biol 2012; 795: 109–18.
Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen 2001; 6: 429–40.
Niesen FH, Berglund H, Vedadi M . The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2007; 2: 2212–21.
Matulis D, Kranz JK, Salemme FR, Todd MJ . Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry (Mosc) 2005; 44: 5258–66.
Senisterra GA, Markin E, Yamazaki K, Hui R, Vedadi M, Awrey DE . Screening for ligands using a generic and high-throughput light-scattering-based assay. J Biomol Screen 2006; 11: 940–8.
Vedadi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty PJ Jr, Wasney GA, et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci U S A 2006; 103: 15835–40.
Holdgate GA, Ward WH . Measurements of binding thermodynamics in drug discovery. Drug Discov Today 2005; 10: 1543–50.
Fedorov O, Marsden B, Pogacic V, Rellos P, Muller S, Bullock AN, et al. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A 2007; 104: 20523–8.
Picaud S, Da Costa D, Thanasopoulou A, Filippakopoulos P, Fish PV, Philpott M, et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET bromodomains. Cancer Res 2013; 73: 3336–46.
Sekiguchi M, Kobashigawa Y, Moriguchi H, Kawasaki M, Yuda M, Teramura T, et al. High-throughput evaluation method for drug association with pregnane X receptor (PXR) using differential scanning fluorometry. J Biomol Screen 2013; 18: 1084–91.
DeSantis K, Reed A, Rahhal R, Reinking J . Use of differential scanning fluorimetry as a high-throughput assay to identify nuclear receptor ligands. Nucl Recept Signal 2012; 10: e002.
Blumberg B, Evans RM . Orphan nuclear receptors — new ligands and new possibilities. Genes Dev 1998; 12: 3149–55.
Wilkinson J, Hallis T, Hermanson S, Bi K . Development and validation of a cell-based assay for the nuclear receptor retinoid-related orphan receptor gamma. Assay Drug Dev Technol 2011; 9: 125–35.
Wilkinson JM, Hayes S, Thompson D, Whitney P, Bi K . Compound profiling using a panel of steroid hormone receptor cell-based assays. J Biomol Screen 2008; 13: 755–65.
Paguio A, Stecha P, Wood KV, Fan F . Improved dual-luciferase reporter assays for nuclear receptors. Curr Chem Genomics 2010; 4: 43–9.
Ou-Yang SS, Lu JY, Kong XQ, Liang ZJ, Luo C, Jiang H . Computational drug discovery. Acta Pharmacol Sin 2012; 33: 1131–40.
Zheng M, Liu X, Xu Y, Li H, Luo C, Jiang H . Computational methods for drug design and discovery: focus on China. Trends Pharmacol Sci 2013; 34: 549–59.
Huang H, Yu Y, Gao Z, Zhang Y, Li C, Xu X, et al. Discovery and optimization of 1,3,4-trisubstituted-pyrazolone derivatives as novel, potent, and nonsteroidal farnesoid X receptor (FXR) selective antagonists. J Med Chem 2012; 55: 7037–53.
Fu J, Si P, Zheng M, Chen L, Shen X, Tang Y, et al. Discovery of new non-steroidal FXR ligands via a virtual screening workflow based on phase shape and induced fit docking. Bioorg Med Chem Lett 2012; 22: 6848–53.
Shen J, Jiang J, Kuang G, Tan C, Liu G, Huang J, et al. Discovery and structure-activity analysis of selective estrogen receptor modulators via similarity-based virtual screening. Eur J Med Chem 2012; 54: 188–96.
Shen J, Tan C, Zhang Y, Li X, Li W, Huang J, et al. Discovery of potent ligands for estrogen receptor beta by structure-based virtual screening. J Med Chem 2010; 53: 5361–5.
Xu T, Wang XH, Zhong B, Nurieva RI, Ding S, Dong C . Ursolic acid suppresses interleukin-17 (IL-17) production by selectively antagonizing the function of ROR lambda t protein. J Biol Chem 2011; 286: 22707–10.
Kumar N, Solt LA, Conkright JJ, Wang YJ, Istrate MA, Busby SA, et al. The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]-benzenesulfonamide] Is a novel retinoic acid receptor-related Orphan receptor-alpha/gamma inverse agonist. Mol Pharmacol 2010; 77: 228–36.
Wang YJ, Kumar N, Nuhant P, Cameron MD, Istrate MA, Roush WR, et al. Identification of SR1078, a synthetic agonist for the orphan nuclear receptors ROR alpha and ROR gamma. ACS Chem Biol 2010; 5: 1029–34.
Kumar N, Lyda B, Chang MR, Lauer JL, Solt LA, Burris TP, et al. Identification of SR2211: a potent synthetic ROR gamma-selective Modulator. ACS Chem Biol 2012; 7: 672–7.
Solt LA, Kumar N, He YJ, Kamenecka TM, Griffin PR, Burris TP . Identification of a selective ROR gamma ligand that suppresses T(H)17 cells and stimulates T regulatory cells. ACS Chem Biol 2012; 7: 1515–9.
Huh JR, Englund EE, Wang H, Huang RL, Huang PX, Rastinejad F, et al. Identification of potent and selective diphenylpropanamide ROR gamma Inhibitors. ACS Med Chem Lett 2013; 4: 79–84.
Khan PM, El-Gendy BEDM, Kumar N, Garcia-Ordonez R, Lin L, Ruiz CH, et al. Small molecule amides as potent ROR-gamma selective modulators. Bioorg Med Chem Lett 2013; 23: 532–6.
Gaweco A, Tilley JW, Walker J, Palmer S, Blinn J . Preparation of quinoline compounds as ROR (retinoid-related orphan receptor) modulators and their therapeutic uses. WO 2013/159095. Apr 22. 2013.
Glick GD, Toogood PL, Romero AG, Vanhuis CA, Aicher TD, Kaub C, et al. Tetrahydroquinoline and related bicyclic compounds for inhibition of ROR activity and the treatment of disease and their preparation. WO 2012/064744. Nov 8. 2012.
Aicher TBK, Lapointe B, Simov V, Stein K, Thomas W, Toogood PvHC, et al. Tetrahydronaphthyridine and related bicyclic compounds for inhibition of RORγ activity and the treatment of disease. WO 2013/169704. Nov 14. 2013.
Barr KJ, Maclean J, Zhang H, Beresis Rt, Zhang D . 3-Cyclohexenyl and cyclohexyl substituted indole and indazole compounds as rorgammat inhibitors and uses thereof. WO2014/028597. Feb 20. 2014.
Zhang W, Zhang J, Fang LP, Zhou L, Wang S, Xiang ZJ, et al. Increasing human Th17 differentiation through activation of orphan nuclear receptor retinoid acid-related orphan receptor gamma (ROR gamma) by a class of aryl amide compounds. Mol Pharmacol 2012; 82: 583–90.
Chaudhari SS, Thomas A, Khairatkar-Joshi N, Bajpai M . Preparation of aryl and heteroaryl amide compounds as RORγt modulator. WO 2013/171729. Aug 29. 2013.
Birault VCAJ, Harrison S, Le J, Sulfonamide, Rabino M . Sulfonamide Compounds and their use in the modulation retinoid-related orphan receptor. WO 2013/045431. Apr 4. 2013.
Birault V, Campbell AJ, Harrison S, Le J, Shukla L . Preparation of heterocyclic benzenesulfonamides as RORγ modulators for treatment of inflammatory, metabolic, or autoimmune disorders. WO 2013/160419. Apr 25. 2013.
Fauber BP, Rene O, Burton B, Everett C, Gobbi A, Hawkins J, et al. Identification of tertiary sulfonamides as RORc inverse agonists. Bioorg Med Chem Lett 2014; 24: 2182–7.
Kotoku M, Maeba T, Seki N, Hirashima S, Fujioka S, Obika S, et al. Preparation of isoxazoles and their use as ROR-γ antagonists and pharmaceuticals. WO 2014/065413. Oct 25. 2014.
Fauber BP, de Leon Boenig G, Burton B, Eidenschenk C, Everett C, Gobbi A, et al. Structure-based design of substituted hexafluoroisopropanol-arylsulfonamides as modulators of RORc. Bioorg Med Chem Lett 2013; 23: 6604–9.
Acknowledgements
We gratefully acknowledge financial support from the National Natural Science Foundation of China (Grant 81373325), the National Key Basic Research Program of China (973 Program, Grant 2013CB910601), the “100 Talents Project” of CAS, and the Guangzhou Bureau of Science and Information Technology, China, (Grants 2012Y2-00051, 2013Y2-00048),“Interdisciplinary Cooperation Team” Program for Science and Technology Innovation of the CAS.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhang, Y., Luo, Xy., Wu, Dh. et al. ROR nuclear receptors: structures, related diseases, and drug discovery. Acta Pharmacol Sin 36, 71–87 (2015). https://doi.org/10.1038/aps.2014.120
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2014.120
Keywords
This article is cited by
-
Liver receptor homolog-1: structures, related diseases, and drug discovery
Acta Pharmacologica Sinica (2024)
-
Ginseng-derived panaxadiol ameliorates STZ-induced type 1 diabetes through inhibiting RORγ/IL-17A axis
Acta Pharmacologica Sinica (2023)
-
T helper cell polarity determines salt sensitivity and hypertension development
Hypertension Research (2023)
-
Single-cell epigenomics and spatiotemporal transcriptomics reveal human cerebellar development
Nature Communications (2023)
-
Unraveling the physiological roles of retinoic acid receptor-related orphan receptor α
Experimental & Molecular Medicine (2021)