Abstract
Aim:
Myeloperoxidase (MPO) and glutathione S-transferase pi 1 (GSTP1) are important carcinogen-metabolizing enzymes. The aim of this study was to investigate the association between the common polymorphisms of MPO and GSTP1 genes and lung cancer risk in Chinese Han population.
Methods:
A total of 266 subjects with lung cancer and 307 controls without personal history of the disease were recruited in this case control study. The tagSNPs approach was used to assess the common polymorphisms of MOP and GSTP1 genes and lung cancer risk according to the disequilibrium information from the HapMap project. The tagSNP rs7208693 was selected as the polymorphism site for MPO, while the haplotype-tagging SNPs rs1695, rs4891, rs762803 and rs749174 were selected as the polymorphism sites for GSTP1. The gene polymorphisms were confirmed using real-time PCR, cloning and sequencing.
Results:
The four GSTP1 haplotype-tagging SNPs rs1695, rs4891, rs762803 and rs749174, but not the MPO tagSNP rs7208693, exhibited an association with lung cancer susceptibility in smokers in the overall population and in the studied subgroups. When Phase 2 software was used to reconstruct the haplotype for GSTP1, the haplotype CACA (rs749174+rs1695 + rs762803+rs4891) exhibited an increased risk of lung cancer among smokers (adjust odds ratio 1.53; 95%CI 1.04–2.25, P=0.033). Furthermore, diplotype analyses demonstrated that the significant association between the risk haplotype and lung cancer. The risk haplotypes co-segregated with one or more biologically functional polymorphisms and corresponded to a recessive inheritance model.
Conclusion:
The common polymorphisms of the GSTP1 gene may be the candidates for SNP markers for lung cancer susceptibility in Chinese Han population.
Similar content being viewed by others
Introduction
The genetic variations of carcinogen-metabolizing enzymes may lead to interindividual differences in the levels of the internal carcinogenic dose and lead to a differential risk in individuals with similar exposures1,2. Phase I enzymes such as myeloperoxidase (MPO) and cytochrome P450 can metabolically activate a wide range of tobacco mutagens to DNA-damaging metabolites3,4. After phase I bioactivation, phase II enzymes (such as glutathione S-transferase) can detoxify the DNA-damaging metabolites via conjugation with endogenous molecules to form hydrophilic conjugates5,6. The accumulated levels of the reactive metabolic intermediates are partly dependent on the metabolic balance of the phase I and II enzymes. Thus, interindividual genetic differences in this metabolic balance may affect an individual's, and especially a smoker's, susceptibility to lung cancer.
MPO is a phase I metabolic enzyme found in neutrophils and monocytes. In experimental systems, it has been shown that MPO can metabolize tobacco smoke procarcinogens into highly reactive intermediates that can damage DNA7. In addition, recent research has shown that MPO may play a major role during pulmonary carcinogenesis, such as the inhibition of nucleotide excision repair in human pulmonary epithelial cells8, DNA damage caused by hypochlorous acid and other reactive oxygen species9 and the methylation of P1610. Clinical and epidemiological studies on the association between the MPO G463A polymorphism and lung cancer susceptibility indicate that the association exists in Caucasian and Chinese populations, although it is not significant11. Because both experimental and epidemiological studies indicated that MPO could play an important role in pulmonary carcinogenesis, the hypothesis that polymorphism(s) of MPO may affect lung cancer risk deserved further study, both independently and at the genome-wide level.
Glutathione S-transferase pi 1 (GSTP1), a member of the GST superfamily, has been identified as the major metabolic GST enzyme in the human lung12. In experimental systems, it has been shown that GSTP1 plays a critical role in the development of lung carcinogenesis following exposure to tobacco-related carcinogens, such as the detoxification of electrophilic diol-epoxides produced by the metabolism of polycyclic aromatic hydrocarbons5,6 and aberrant promoter methylation of the P16 tumor suppressor gene and the O(6)-methylguanine-DNA methyltransferase (MGMT) DNA repair gene13,14. However, some epidemiological studies demonstrating the association between polymorphisms of GSTP1 and lung cancer risk yielded inconclusive results15,16,17,18,19,20,21. Thus, further study of GSTP1 as a candidate gene in lung cancer risk alone or at the genome-wide level was necessary.
The International HapMap project (www.hapmap.org) is the most important post-genomic project that is able to provide relatively exact SNP information for the Chinese population and provide a systematic framework of linkage disequilibrium (LD) and haplotype structure for SNPs22. After considering the common disease–common variant hypothesis, association analysis using LD mapping would seem to be a reasonable approach in narrowing down the number of potential risk genes or variants for the disease. It is conceivable that the tagging SNPs for GSTP1 and MPO may be used to test the association between polymorphisms and susceptibility.
The HapMap project had just completed collecting phase II data for the Chinese population when we initiated our study in November 2008 (HapMap Data Rel#24/phase II on NCBI B36 assembly, dbSNP b126). The HapMap project provided 6 common SNPs for the GSTP1 gene and 1 common SNP for MPO (frequency >5%) and the linkage disequilibrium information for the Chinese Han population. In our study, we aimed to evaluate the association between the common variants in GSTP1 and MPO and the susceptibility to lung cancer using the tagSNPs approach and integer analyses.
Materials and methods
Study population and data collection
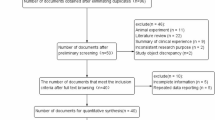
This research was based on a case control study on lung cancer conducted at the Tianjin Medical University General Hospital and Tianjin Key Laboratory of Lung Cancer Metastasis and Tumor Microenvironment, Tianjin Lung Cancer Institute, China. Patients that were diagnosed histologically and cytologically with lung cancer and a comparable group of hospital-based control subjects without the disease were recruited from February 2008 and October 2009. The information from a total of 266 patients with lung cancer and 307 controls was collected. The cases included 120 (45.1%) squamous cell carcinomas (Scc), 99 (37.2%) adenocarcinomas (Ad), 23 (16.9%) small-cell lung carcinomas (SCLC) and 24 (9.0%) other lung carcinomas. The control subjects were randomly selected from a pool of healthy volunteers who had visited the General Health Check-up Center at Tianjin Medical University General Hospital during the same period. The cases and controls were all ethnic Han Chinese.
A detailed questionnaire was completed for each patient and each control by a trained interviewer. The questionnaire included information on their gender, age, tobacco smoking history, tumor histotype, environmental exposure and diet. For the smoking status of the subjects, a person who had smoked at least 100 cigarettes during his or her lifetime was considered to be a smoker23. The cumulative cigarette dose (pack-years) was calculated using the following formula: pack-years=(packs per day)×(years smoked). We further categorized the subjects as never smokers, light smokers (≤27 pack-years, the mean of the pack-year) and heavy smokers (>27 pack-years) according to their smoking status. Informed consent was obtained from all participants, and the study was approved by the Research Ethics Committee of the Tianjin Medical University General Hospital.
Selection of haplotype-based tagSNPs
Genotypes for SNPs in GSTP1 representing the Han Chinese were downloaded from the HapMap database (http://www.hapmap.org, HapMap Data Rel#24/phase II on NCBI B36 assembly, dbSNP b126). LD and haplotypes in the GSTP1 gene were determined using the Haploview 4.1 software (Broad Institute of MIT and Harvard, Cambridge, MA, USA) at default values. LD was estimated between the pairs of SNPs using the D-statistic. The haplotype block structure was determined using the confidence interval option. Four tagSNPs, rs1695, rs4891, rs762803, and rs749174, were chosen to capture the common variants within GSTP1; these four SNPs had also been validated by other submissions (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?locusId=2950). In the HapMap Data Rel#24, there is only 1 common SNP for MPO, rs2071409 (in intron). However, in the dbSNP, there are three common SNPs - rs7208693 (in the 53 acid position, the allele T→G could cause a phe→val change), rs35921530 (in intron) and rs2856857 (in intron). Hence, we chose rs7208693 to capture the common variants within MPO (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=7208693).
DNA extraction
Genomic DNA was isolated from peripheral blood using a standard kit-based method (Axygen). The DNA concentration was adjusted to 40 mmol/L with TE buffer, and all DNA preparations were stored at -20 °C until they were used for genotyping.
Genotyping
A TaqMan® (ABI) genotyping assay was employed to genotype all samples for the 5 selected tagSNPs. For each of the SNPs, primer-probe sets (Table 1) were designed using the Applied Biosystems design service (Applied Biosystems, Foster City, CA, USA). We carried out real-time PCR on 10 ng of genomic DNA using the TaqMan® universal PCR master mix (Applied Biosystems), forward and reverse primers and FAM- and VIC-labeled probes. Real-time PCR was performed using 5.0 μL of the universal master mix (Applied Biosystems), 0.25 μL of the primer-probe mix, 2.25 μL of RNase- and DNase-free water and 2.5 μL of DNA (40 mmol/L). The assay conditions were as follows: 10 min at 95 °C and 40 cycles of 15 s at 92 °C and 1 min at 58 °C. The real-time PCR 7500 system (Applied Biosystems; SDS version 1.4 software) was used to perform and analyze the genotyping. For the purposes of quality control, more than 2 negative controls containing all reagents but substituting water for DNA were included in each amplification set. The genotyping was carried out blinded to the case control status. A 10% random sample was repeated to verify the genotyping results. Each genotype of the 5 SNPs was cloned and sequenced randomly.
Statistical analysis
Demographic and clinical information between the cases and controls was compared using chi-square tests for categorical variables and Student's t-test for continuous variables, where appropriate. The Hardy–Weinberg equilibrium was confirmed by chi-square analysis. The departure of genotype frequencies from those expected under the Hardy-Weinberg equilibrium was assessed among Chinese controls by the asymptotic Pearson's chi-square test with one degree of freedom. The estimated GSTP1 haplotypes for the 266 cases and 307 controls were determined using the PHASE Version 2 software (http://www.stat.washington.edu/stephens/software.html), which uses a Bayesian algorithm for haplotype reconstruction. The best-pairs diplotypes were estimated for each individual, and the diplotypes for each individual were split into individual haplotypes. The diplotypes with the risk haplotype were designated as the at-risk diplotypes, while the other diplotypes were combined and designated as the non-at-risk diplotypes. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using a logistic regression analysis where the log odds of lung cancer were adjusted for smoking (as a categorical variable), age (as a continuous variable) and sex (as a categorical variable). To detect important differences in the population subgroups, stratification by a subgroup analysis of clinically relevant factors (eg, smoking status and histological types) was performed. All tests were two-sided, and a P value of 0.05 or less was considered significant. All analyses were performed using SPSS (Statistical Package for the Social Sciences) software, version 11.5 (Chicago, USA).
Results
General characteristics of the included subjects
The demographics of the cases and controls enrolled in this study are shown in Table 2. There were no significant differences between the cases and controls in terms of the mean age or gender distribution, suggesting that the matching based on these two variables was adequate. The case group had a higher prevalence of smokers than the control group (P<0.01) and the smoking index of pack-years for the smokers in lung cancer group was significantly higher than that in the control group (P<0.01). These differences were adjusted using multivariate analysis.
Smoking and lung cancer risk
After adjustments for age at diagnosis, gender and smoking history, a statistically increased lung cancer risk (OR=3.30, 95% CI 2.18-4.99, P<0.01) was confirmed for the cases when compared to the never smokers. The further stratified analysis according to smoking status indicated that the association appeared mainly in the heavy smokers subgroup (>27 pack-years) (OR=8.98, 95% CI 5.40–14.96, P<0.01). Stratified by histological type, tobacco smoking had a significantly increased lung cancer risk (OR=8.28, 95% CI 4.24–16.20, P<0.01), mainly in the squamous cell carcinoma subgroup (Table 3). Furthermore, a strong and significant dose–response relationship between lung cancer risk and the pack-years of smoking (P<0.01) was found, especially in the squamous carcinoma subgroup.
Genetic polymorphisms of MPO and GSTP1 and lung cancer susceptibility
The distribution of the rs1695 (A/G), rs4891 (A/G), rs762803 (C/A), rs749174 (C/T), and rs7208693 (C/A) genotypes among the cases and controls is shown in Table 4. The genotype distributions of the 5 SNPs in the 573 subjects are consistent with the data from Phase 3 of the HapMap project (by the time our genotyping was completed, the HapMap data had been updated to Phase 3). The genotype distributions of the 5 polymorphisms among the controls were in Hardy-Weinberg equilibrium. The distribution of the rs7208693 (C/A) genotype of MPO was not different between the cases and controls, not only in the overall population but also in the subgroups stratified according to smoking and histological type. The distributions of the rs1695 (A/G), rs4891 (A/G), rs762803 (C/A), and rs749174 (C/T) genotypes of GSTP1 were different between the cases and controls in the overall population, especially in the smoking and squamous cell carcinoma subgroups. In general, no significant association between the four genotypes of GSTP1 and lung cancer risk was found when adjusted for age at diagnosis (continuous), gender (male and female) or smoking status (no smoking, light smoking, heavy smoking) (Table 4). However, in the subgroup analysis, the G allele of rs1695 (A/G), G allele of rs4891 (A/G), A allele of rs762803 (C/A) and T allele of rs749174 (C/T) statistically decreased the lung cancer susceptibility in the smoking and squamous cell carcinoma patients (P<0.05). The result is concordant with the conclusion that tobacco smoking is the major risk factor for squamous cell carcinoma.
Phase 2 software was used to reconstruct the haplotypes of the rs1695 (A/G), rs4891 (A/G), rs762803 (C/A), and rs749174 (C/T) loci of GSTP1, and 13 out of the possible 16 (24) haplotypes were observed. For statistical advantage, 11 haplotypes with a rare frequency (<2%) were combined with others for further analysis (data not shown). Table 5 shows the inferred haplotype distributions for the cases and controls as well as the lung cancer risk according to haplotype. The haplotype CACA (rs749174+rs1695 + rs762803+rs4891) was associated with an increased risk of lung cancer (adjusted OR=1.39, 95% CI 1.01–1.91, for the overall population; adjusted OR=1.53, 95% CI 1.04–2.25, for the smoker sub-population). Because diplotype analysis showed that the diplotype with 0 or 1 copies of CACA decreased the risk of lung cancer, we selected the combined diplotypes with 0 or 1 copies of CACA as the reference against which to analyze the OR. Diplotype analysis showed that the diplotype with 2 copies of CACA increased the lung cancer risk in the overall population and in the smoker and squamous cell carcinoma sub-populations (Table 6).
Discussion
China is experiencing rapid industrialization, and lung cancer incidence has increased over the years and become the most common type of malignant tumor, especially in some metropolises such as Tianjin and Shanghai24,25. In addition, further attention needs to be paid to new epidemiological factors, such as an increase in the number of female cases and adenocarcinoma subtypes, as these may suggest that a new etiological factor or a previously unknown factor is influencing the incidence of lung cancer26.
The MPO gene is located on chromosome 17q23.1 containing 12 exons and 11 introns. When we initiated the present study in November 2008, the HapMap project had just been completed for phase II data. Therefore, we retrieved SNP data from the HapMap database and found 1 common SNP (rs2071409, in an intron). However, the dbSNP contains 3 common SNPs– rs7208693 (in the 53 acid position, the allele T→G could cause a phe→val change), rs35921530 (in intron) and rs2856857 (in intron). In addition to the tagSNP approach, a previously utilized method was to investigate functional SNPs in coding regions. Moreover, the HapMap project was ongoing and the data were continuously updated. Hence, we chose the rs7208693 SNP to capture the common variants within MPO. In our study, the allelic distribution of 7208693(A/C) was 0.91 for the C allele and 0.9 for the A allele, which is consistent with the dbSNP and HapMap phase 3 data (rs7208693 was reported as a tagger in the updated phase 3 data in February 2009). In the updated HapMap 3 data, rs7208693 and rs2071409 (in intron 7) are listed as common SNPs and are in complete LD. We can conclude that the common variants of MPO are not associated with lung cancer risk. In addition, the percentage of the TT genotype for MPO is only 1.4%, which cannot be used to evaluate an association between polymorphisms and lung cancer susceptibility.
There is another common SNP of the MPO gene, rs2333227, located in the promoter region 463 base pairs upstream. The possible mechanism by which rs2333227 may affect lung cancer risk is to decrease MPO gene expression by destroying a transcript-binding site. Rs2333227 has been investigated extensively for the association with lung cancer risk, and a recent pooled-analysis indicated that the association did exist, although with weak significance11,27,28,29,30,31,32,33. Hence, we did not choose this SNP for the present study. We also evaluated the relationship between the myeloperoxidase G463A genetic polymorphism and lung cancer susceptibility by meta-analysis, and the results indicated that the polymorphism was not significantly associated with lung cancer risk in the Caucasian or East Asian population34.
The GSTP1 gene, mapped to chromosome 11q13 and located in a 2.8 kb region, consists of 7 exons and 6 introns. In the experimental system, it has been proven that GSTP1 plays an important role in the metabolism of lung carcinogens and DNA damage13,35,36. Singh et al37 examined 230 subjects, including 115 workers occupationally exposed to organophosphate pesticides, to determine the association between genetic polymorphisms of GSTP1 and susceptibility to DNA damage. The results showed that GSTP1 polymorphisms might be related to interindividual differences in DNA damage arising from the gene-environment interactions in occupationally exposed workers37.
The rs1695 (Ile105Val, A/G) polymorphism of GSTP1 has been investigated extensively, and the results were not conclusive16,38,39,40,41,42. We also evaluated the relationship between the GSTP1 Ile105Val (rs1695) genetic polymorphism and lung cancer susceptibility by meta-analysis, and the results indicated that the polymorphism was not significantly associated with lung cancer risk in the Caucasian or East Asian populations or with gender, smoking status or histological type subgroups. The meta-analysis did not research the association in the subgroups based on the amount smoked, as some studies were not able to provide the number of pack-years of cigarette smoking43. Cote performed a pooled-analysis to investigate the association and found that the G allele of rs1695 decreased the risk of lung cancer in Caucasian heavy-smokers (>48 pack-years) and that there was a statistically significant trend between the amount smoked and the risk of lung cancer; there was an increased risk in the adenocarcinoma and non-smoker subgroups in the Asian population15. We reviewed the studies adopted in the pooled analysis and found that the sample size of the Asian population (adenocarcinoma subgroup) was only 19541,44,45,46. Our study indicates that the G allele of rs1695 decreased lung cancer risk in Chinese smokers, which is consistent with the conclusion for the Caucasian population in the Cote study.
If we select haplotype CACA (rs749174+rs1695+rs762803+ rs4891) as the risk factor, it corresponds to a recessive inheritance model. According to the our literature review, a reasonable explanation for the decreased lung cancer risk due to the G allele of rs1695 is that the G allele of rs1695 results in reduced GSTP1 enzymatic activity in the cell47. However, the exact mechanism by which the other three SNPs of GSTP1 decrease the lung cancer risk in the smoking or Scc subgroups was not clear. GSTP1 belongs to a family of enzymes that play an important role in detoxification by catalyzing the conjugation of many hydrophobic and electrophilic compounds with reduced glutathione. GSTP1 is also known as a polymorphic gene encoding active, functionally different GSTP1 variant proteins that are thought to function in xenobiotic metabolism and play a role in susceptibility to cancer and other diseases. Nevertheless, these characterizations, while important for developing a hypothesis regarding the biologic mechanisms through which carcinogenesis evolves, do not necessarily represent what is occurring in the environment of the human lung.
Timofeeva et al investigated the association between the rs1695, rs947895 (+991C>A) and rs4891 genetic polymorphisms of GSTP1 and early onset lung cancer48. Their study included 638 Caucasian patients under the age of 51 with confirmed primary lung cancer and 1300 cancer-free control individuals, matched by age and sex. Their results revealed that no significant association was found for any of the analyzed polymorphisms and overall lung cancer risk, although their subgroup analysis revealed smoking-specific effects48. The latest study of a Turkish population showed that the GSTP1 exon 6 polymorphism (rs4891) may be an important factor in determining lung cancer susceptibility49.
There is no risk genotype or allele for rs7208693, so it is difficult to evaluate the combination of GSTP1 and MPO with risk. We attempted to analyze the association by selecting the combination of the CC genotype of rs7208693 and 0 or 1 copies of CACA as the reference and investigating the risk caused by the combination of the AC genotype of rs7208693 and 2 copies of CACA. This resulted in an OR of 1.64 (95% CI, 0.86–3.12, P=0.134) in the overall population and 1.84 (95% CI, 0.81–4.2, P=0.148) in the smoker population. This was no different from the OR caused by a single GSTP1 polymorphism. Hence, in our opinion, there is no combined contribution of the common polymorphisms of GSTP1 and MPO.
There are several limitations to this study. First, as this is a hospital-based case control study, our control group was not ideal. Second, although the results had been adjusted for smoking variables in our analysis, variables such as exposure to secondhand smoke, diet and exposure to environmental and occupational factors were not adjusted in our logistic regression models because of incomplete and missing information. It is likely that if the confounders involved in the smoking populations were controlled, the ORs would increase significantly. The third limitation was our low statistical power for the sample size of our current study. Although the sample size of our study met the requirement according to the SNPs-selecting standard (MAF>5%), it was still low enough to restrict the significance. Finally, using the tagSNP approach may have resulted in the loss of important SNP information, such as the SNPs (MAF<5%) and the untyped SNPs.
In conclusion, our study investigated the association between common MPO and GSTP1 polymorphisms and lung cancer risk using the tagSNP approach, which may reflect extensive information regarding the candidate genes. Our study suggested that the common polymorphisms of GSTP1 could be candidate SNP markers for lung cancer susceptibility in the Chinese population in future GWAS studies.
Author contribution
Qing-hua ZHOU designed the research; Jun-dong GU, Feng HUA, and De-jie ZHENG recruited research subjects; Jun-dong GU, Feng HUA, and Chao-rong MEI performed the research; Guo-fan WANG contributed new analytic tools; Feng HUA analyzed the data; and Feng HUA and Jun-dong GU wrote the paper.
References
Gresner P, Gromadzinska J, Wasowicz W . Polymorphism of selected enzymes involved in detoxification and biotransformation in relation to lung cancer. Lung Cancer 2007; 57: 1–25.
Li XF, Zhang J, Su C, Zhao X, Tang L, Zhou C . The association between polymorphisms in the DNA nucleotide excision repair genes and RRM1 gene and lung cancer risk. Thoracic Cancer 2012; 3: 239–48.
Zhang X, D'Agostino J, Wu H, Zhang QY, von WL, Murphy SE, et al. CYP2A13: variable expression and role in human lung microsomal metabolic activation of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. J Pharmacol Exp Ther 2007; 323: 570–8.
Knaapen AM, Gungor N, Schins RP, Borm PJ, Van Schooten FJ . Neutrophils and respiratory tract DNA damage and mutagenesis: a review. Mutagenesis 2006; 21: 225–36.
Ritchie KJ, Henderson CJ, Wang XJ, Vassieva O, Carrie D, Farmer PB, et al. Glutathione transferase pi plays a critical role in the development of lung carcinogenesis following exposure to tobacco-related carcinogens and urethane. Cancer Res 2007; 67: 9248–57.
Constantin D, Mehrotra K, Rahimtula A, Moldeus P, Jernstrom B . Stimulatory effects of sulfur and nitrogen oxides on carcinogen activation in human polymorphonuclear leukocytes. Environ Health Perspect 1994; 102: 161–4.
Arlt VM, Schmeiser HH, Osborne MR, Kawanishi M, Kanno T, Yagi T, et al. Identification of three major DNA adducts formed by the carcinogenic air pollutant 3-nitrobenzanthrone in rat lung at the C8 and N2 position of guanine and at the N6 position of adenine. Int J Cancer 2006; 118: 2139–46.
Gungor N, Godschalk RW, Pachen DM, Van Schooten FJ, Knaapen AM . Activated neutrophils inhibit nucleotide excision repair in human pulmonary epithelial cells: role of myeloperoxidase. FASEB J 2007; 21: 2359–67.
Gungor N, Knaapen AM, Munnia A, Peluso M, Haenen GR, Chiu RK, et al. Genotoxic effects of neutrophils and hypochlorous acid. Mutagenesis 2010; 25: 149–54.
Chan EC, Lam SY, Fu KH, Kwong YL . Polymorphisms of the GSTM1, GSTP1, MPO, XRCC1, and NQO1 genes in Chinese patients with non-small cell lung cancers: relationship with aberrant promoter methylation of the CDKN2A and RARB genes. Cancer Genet Cytogenet 2005; 162: 10–20.
Taioli E, Benhamou S, Bouchardy C, Cascorbi I, Cajas-Salazar N, Dally H, et al. Myeloperoxidase G-463A polymorphism and lung cancer: a HuGE genetic susceptibility to environmental carcinogens pooled analysis. Genet Med 2007; 9: 67–73.
Spivack SD, Hurteau GJ, Fasco MJ, Kaminsky LS . Phase I and II carcinogen metabolism gene expression in human lung tissue and tumors. Clin Cancer Res 2003; 9: 6002–11.
Gilliland FD, Harms HJ, Crowell RE, Li YF, Willink R, Belinsky SA . Glutathione S-transferase P1 and NADPH quinone oxidoreductase polymorphisms are associated with aberrant promoter methylation of P16(INK4a) and O(6)-methylguanine-DNA methyltransferase in sputum. Cancer Res 2002; 62: 2248–52.
Wlodarczyk M, Nowicka G . Common polymorphisms in CYP1A1, GSTM1, GSTT1, GSTP1, and XPD genes and endogenous DNA damage. Mol Biol Rep 2012; 39: 5699–704.
Cote ML, Chen W, Smith DW, Benhamou S, Bouchardy C, Butkiewicz D, et al. Meta- and pooled analysis of GSTP1 polymorphism and lung cancer: a HuGE-GSEC review. Am J Epidemiol 2009; 169: 802–14.
Miller DP, Asomaning K, Liu G, Wain JC, Lynch TJ, Neuberg D, et al. An association between glutathione S-transferase P1 gene polymorphism and younger age at onset of lung carcinoma. Cancer 2006; 107: 1570–7.
To-Figueras J, Gene M, Gomez-Catalan J, Pique E, Borrego N, Corbella J . Lung cancer susceptibility in relation to combined polymorphisms of microsomal epoxide hydrolase and glutathione S-transferase P1. Cancer Lett 2001; 173: 155–62.
Cote ML, Yoo W, Wenzlaff AS, Prysak GM, Santer SK, Claeys GB, et al. Tobacco and estrogen metabolic polymorphisms and risk of non-small cell lung cancer in women. Carcinogenesis 2009; 30: 626–35.
Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland LB, et al. A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of non-small cell lung cancer in smokers. Carcinogenesis 2008; 29: 1164–9.
Larsen JE, Colosimo ML, Yang IA, Bowman R, Zimmerman PV, Fong KM . CYP1A1 Ile462Val and MPO G-463A interact to increase risk of adenocarcinoma but not squamous cell carcinoma of the lung. Carcinogenesis 2006; 27: 525–32.
Stucker I, Hirvonen A, de Waziers I, Cabelguenne A, Mitrunen K, Cenee S, et al. Genetic polymorphisms of glutathione S-transferases as modulators of lung cancer susceptibility. Carcinogenesis 2002; 23: 1475–81.
Phillips C . Online resources for SNP analysis: a review and route map. Mol Biotechnol 2007; 35: 65–97.
Hernandez JL, Cunado A, Gonzalez-Macias J . Knowledge of the packs/year as an index of the smoking habit among the clinicians of an Internal Medicine Department. Med Clin (Barc) 2003; 121: 359.
Chang S, Dai M, Ren JS, Chen YH, Guo LW . Estimates and prediction on incidence, mortality and prevalence of lung cancer in China in 2008. Zhonghua Liu Xing Bing Xue Za Zhi 2012; 33: 391–4.
Han X, Qiao P, Xie M, Zhao J, Huang CX, Zhang HW, et al. The incidence and mortality of lung cancer among residents in Yangpu district of Shanghai from 2002 to 2010. Zhonghua Zhong Liu Za Zhi 2012; 34: 712–8.
Kligerman S, White C . Epidemiology of lung cancer in women: risk factors, survival, and screening. AJR Am J Roentgenol 2011; 196: 287–95.
Wu XM, Zhou YK, Ren S, Hao QL . Gene polymorphism of myeloperoxidase and genetic susceptibility to lung cancer. Ai Zheng 2003; 22: 912–5.
Lu W, Qi J, Xing D, Tan W, Miao X, Su W, et al. Lung cancer risk associated with genetic polymorphism in myeloperoxidase (-463 G/A) in a Chinese population. Zhonghua Zhong Liu Za Zhi 2002; 24: 250–3.
Feyler A, Voho A, Bouchardy C, Kuokkanen K, Dayer P, Hirvonen A, et al. Point: myeloperoxidase -463G → a polymorphism and lung cancer risk. Cancer Epidemiol Biomarkers Prev 2002; 11: 1550–4.
Schabath MB, Spitz MR, Hong WK, Delclos GL, Reynolds WF, Gunn GB, et al. A myeloperoxidase polymorphism associated with reduced risk of lung cancer. Lung Cancer 2002; 37: 35–40.
Kantarci OH, Lesnick TG, Yang P, Meyer RL, Hebrink DD, McMurray CT, et al. Myeloperoxidase -463 (G→A) polymorphism associated with lower risk of lung cancer. Mayo Clin Proc 2002; 77: 17–22.
Yang M, Choi Y, Hwangbo B, Lee JS . Combined effects of genetic polymorphisms in six selected genes on lung cancer susceptibility. Lung Cancer 2007; 57: 135–42.
Le ML, Seifried A, Lum A, Wilkens LR . Association of the myeloperoxidase -463G→a polymorphism with lung cancer risk. Cancer Epidemiol Biomarkers Prev 2000; 9: 181–4.
Hua F, Wang J, Gu J, Li S, Liu H, Zhou Q . A meta analysis on the relationship between myeloperoxidase G-463A genetic polymorphisms and lung cancer susceptibility. Zhongguo Fei Ai Za Zhi 2010; 13: 122–7.
Perera FP, Mooney LA, Stampfer M, Phillips DH, Bell DA, Rundle A, et al. Associations between carcinogen-DNA damage, glutathione S-transferase genotypes, and risk of lung cancer in the prospective Physicians' Health Cohort Study. Carcinogenesis 2002; 23: 1641–6.
Ryberg D, Skaug V, Hewer A, Phillips DH, Harries LW, Wolf CR, et al. Genotypes of glutathione transferase M1 and P1 and their significance for lung DNA adduct levels and cancer risk. Carcinogenesis 1997; 18: 1285–9.
Singh S, Kumar V, Singh P, Thakur S, Banerjee BD, Rautela RS, et al. Genetic polymorphisms of GSTM1, GSTT1 and GSTP1 and susceptibility to DNA damage in workers occupationally exposed to organophosphate pesticides. Mutat Res 2011; 725: 36–42.
Sobti RC, Kaur P, Kaur S, Janmeja AK, Jindal SK, Kishan J, et al. Combined effect of GSTM1, GSTT1 and GSTP1 polymorphisms on histological subtypes of lung cancer. Biomarkers 2008; 13: 282–95.
To-Figueras J, Gene M, Gomez-Catalan J, Pique E, Borrego N, Carrasco JL, et al. Genetic polymorphism of glutathione S-transferase P1 gene and lung cancer risk. Cancer Causes Control 1999; 10: 65–70.
Schneider J, Bernges U, Philipp M, Woitowitz HJ . GSTM1, GSTT1, and GSTP1 polymorphism and lung cancer risk in relation to tobacco smoking. Cancer Lett 2004; 208: 65–74.
Wang J, Deng Y, Cheng J, Ding J, Tokudome S . GST genetic polymorphisms and lung adenocarcinoma susceptibility in a Chinese population. Cancer Lett 2003; 201: 185–93.
Sorensen M, Raaschou-Nielsen O, Brasch-Andersen C, Tjonneland A, Overvad K, Autrup H . Interactions between GSTM1, GSTT1 and GSTP1 polymorphisms and smoking and intake of fruit and vegetables in relation to lung cancer. Lung Cancer 2007; 55: 137–44.
Ma W, Zhang B, Ha F . A meta analysis on relationship between GSTP1 Ile105Val genetic polymorphisms and lung cancer risk. Cancer Res Prevention Treatment 2011; 11: 1311–5.
Liang G, Pu Y, Yin L . Rapid detection of single nucleotide polymorphisms related with lung cancer susceptibility of Chinese population. Cancer Lett 2005; 223: 265–74.
Lin P, Hsueh YM, Ko JL, Liang YF, Tsai KJ, Chen CY . Analysis of NQO1, GSTP1, and MnSOD genetic polymorphisms on lung cancer risk in Taiwan. Lung Cancer 2003; 40: 123–9.
Kiyohara C, Yamamura KI, Nakanishi Y, Takayama K, Hara N . Polymorphism in GSTM1, GSTT1, and GSTP1 and susceptibility to lung cancer in a Japanese population. Asian Pac J Cancer Prev 2000; 1: 293–8.
Sundberg K, Johansson AS, Stenberg G, Widersten M, Seidel A, Mannervik B, et al. Differences in the catalytic efficiencies of allelic variants of glutathione transferase P1-1 towards carcinogenic diol epoxides of polycyclic aromatic hydrocarbons. Carcinogenesis 1998; 19: 433–6.
Timofeeva M, Kropp S, Sauter W, Beckmann L, Rosenberger A, Illig T, et al. Genetic polymorphisms of MPO, GSTT1, GSTM1, GSTP1, EPHX1 and NQO1 as risk factors of early-onset lung cancer. Int J Cancer 2010; 127: 1547–61.
Ada AO, Kunak SC, Hancer F, Soydas E, Alpar S, Gulhan M, et al. Association between GSTM1, GSTT1, and GSTP1 polymorphisms and lung cancer risk in a Turkish population. Mol Biol Rep 2012; 39: 5985–93.
Acknowledgements
The authors thank the physicians, surgeons and pathologists at Tianjin Medical University General Hospital who endorsed the project and the patients and donors who participated in this study. This work was supported by the National “Eleventh Five-Year” Scientific and Technological Support Projects (No 2006BAI02A01).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gu, Jd., Hua, F., Mei, Cr. et al. HapMap-based study on the association between MPO and GSTP1 gene polymorphisms and lung cancer susceptibility in Chinese Han population. Acta Pharmacol Sin 35, 636–644 (2014). https://doi.org/10.1038/aps.2014.11
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2014.11