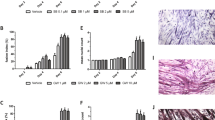
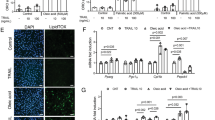
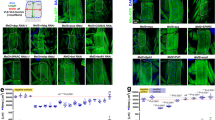
Abstract
Foxo-1, a member of the Foxo forkhead type transcription factors, is markedly upregulated in skeletal muscle in energy-deprived states such as fasting, cancer and severe diabetes. In this study, we target the Foxo-1 mRNA in a mouse skeletal myoblast cell line C2C12 and in vivo models of normal and cancer cachexia mice by a Foxo-1 specific RNA oligonucleotide. Our results demonstrate that the RNA oligonucleotide can reduce the expression of Foxo-1 in cells and in normal and cachectic mice, leading to an increase in skeletal muscle mass of the mice. In search for the possible downstream target genes of Foxo-1, we show that when Foxo-1 expression is blocked both in cells and in mice, the level of MyoD, a myogenic factor, is increased while a muscle negative regulator GDF-8 or myostatin is suppressed. Taken together, these results show that Foxo-1 pays a critical role in development of muscle atrophy, and suggest that Foxo-1 is a potential molecular target for treatment of muscle wasting conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Lynch GS, Schertzer JD, Ryall JG . Therapeutic approaches for muscle wasting disorders. Pharmacol Ther 2007; 113: 461–872.
Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, Emanuel BS et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet 1993; 5: 230–235.
Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, downregulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem 2004; 279: 41114–41123.
Bois PR, Grosveld GC . FKHR (FOXO1a) is required for myotube fusion of primary mouse myoblasts. EMBO J 2003; 22: 1147–1157.
Dijkers PF, Medema RH, Pals C, Banerji L, Thomas NSB, Lam EWF et al. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1). Mol Cell Biol 2000; 20: 9138–9148.
Medema RH, Kops GJ, Bos JL, Burgering BMT . AFX-like forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 2000; 404: 782–787.
Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS et al. Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell 1999; 96: 857–868.
Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KWA, Coffer PJ et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 2002; 419: 316–321.
Grobet L, Pirottin D, Farnir F . Modulating skeletal muscle mass by postnatal, muscle-specific inactivation of the GDF-8 gene. Genesis 2003; 35: 227–238.
Lee SJ, McPherron AC . 2001) Regulation of GDF-8 activity and muscle growth. Proc Natl Acad Sci USA 2001; 98: 9306–9311.
Reisz-Porszasz S, Bhasin S, Artaza JN, Shen R, Sinha-Hikim I, Hogue A et al. Lower skeletal muscle mass in male transgenic mice with muscle-specific overexpression of GDF-8. Am J Physiol Endocrinol Metab 2003; 285: E876–E888.
Wehling M, Cai B, Tidball JG . Modulation of GDF-8 expression during modified muscle use. FASEB J 2000; 14: 103–110.
Baracos VE, DeVivo C, Hoyle DH, Goldberg AL . Activation of the ATP-ubiquitin-proteasome pathway in skeletal muscle of cachectic rats bearing a hepatoma. Am J Physiol 1995; 268: E996–E1006.
Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004; 117: 399–412.
Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore L, Ahima RS et al. Functional improvement of dystrophic muscle by GDF-8 blockade. Nature 2002; 28: 418–421.
Joulia D, Bernardi H, Garandel V, Rabenoelina F, Vernus B, Cabello G . Mechanisms involved in the inhibition of myoblast proliferation and differentiation by GDF-8. Exp Cell Res 2003; 286: 263–275.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Liu, CM., Yang, Z., Liu, CW. et al. Effect of RNA oligonucleotide targeting Foxo-1 on muscle growth in normal and cancer cachexia mice. Cancer Gene Ther 14, 945–952 (2007). https://doi.org/10.1038/sj.cgt.7701091
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cgt.7701091
Keywords
This article is cited by
-
Molecular mechanisms of exercise contributing to tissue regeneration
Signal Transduction and Targeted Therapy (2022)
-
Molecular pathways leading to loss of skeletal muscle mass in cancer cachexia – can findings from animal models be translated to humans?
BMC Cancer (2016)
-
Comparative transcriptome profiling of longissimus muscle tissues from Qianhua Mutton Merino and Small Tail Han sheep
Scientific Reports (2016)
-
Postsurgical Acute Phase Reaction is Associated with Decreased Levels of Circulating Myostatin
Inflammation (2015)
-
Genome-wide identification of FoxO-dependent gene networks in skeletal muscle during C26 cancer cachexia
BMC Cancer (2014)