Abstract
Recent fragmentation of populations as well as historical postglacial recolonization may have significantly affected the population genetic diversity of temperate plant species. Regional allozymic variability was measured at seven loci within and among 12 populations of Calluna vulgaris in the previously glaciated region of Scotland. These results were compared with existing data on south-western continental populations. Low genetic differentiation (FST = 0.024) and lack of consistent geographical pattern were found at the regional level among Scottish populations, implying a high rate of gene flow (Nm = 10.2), probably favoured by the nearly continuous range of C. vulgaris across Scotland and characteristics of the Scottish environment. Scottish populations possessed lower mean allozymic diversity (PLP = 40.48, A = 1.95, He = 0.133) than populations from all the continental regions investigated previously. Belgian populations were genetically more closely related to Scottish than to other continental populations. These last two findings are interpreted with regard to the evolutionary history of the species revealed by palynological data.
Similar content being viewed by others
Introduction
Levels of allozyme variation in plant species are highly variable (Hamrick & Godt, 1989). Life-history traits, such as breeding system, geographical distribution and dispersal ability, explain a significant part of the variability of genetic diversity among plant species (Hamrick & Godt, 1989), but recent and past evolutionary history may also influence genetic diversity and genetic differentiation within plant species. In recent centuries, human activity has led to the fragmentation and isolation of the populations of many temperate plant species. Both theory and experimental measurements show that processes that erode genetic variation, such as genetic drift, are more likely to affect small geographically isolated populations (Young et al., 1996). This loss of genetic variation has traditionally been considered to decrease both the short-term and the long-term adaptability of populations in variable and changing environments (Young et al., 1996). The long-term evolutionary history of temperate plant species, including shifts in distribution, fragmentation and population isolation during and after the last glacial maximum, also undoubtedly greatly affected their population genetic structure. Range expansion following the late-glacial maximum (18000 BP) most probably resulted in a series of bottlenecks for the colonizing populations that should have lead to a loss of alleles and a tendency to homozygosity (Nei et al., 1975; Hewitt, 1996). Also, strong genetic differentiation among groups of populations may arise because of the fragmentation of a species range into geographically isolated refugia during glacial periods (Hewitt, 1996).
Calluna vulgaris (L.) Hull (heather, ling) (Ericaceae) is a low shrub distributed throughout Europe, from Spain to northern Scandinavia and from the Azores to the Urals, with its principal range within western central Europe. Calluna vulgaris may have survived in two regions during the last glaciation events (Weichselian/Wurm): south Continental Europe (including western Spain, the Pyrenees at low and mid-altitude and the Massif Central, France) and southern Britain (Jalut et al., 1982; Huntley & Birks, 1983; Mardonnes & Jalut, 1983; Reille & De Beaulieu, 1988; De Beaulieu & Reille, 1992). Today, C. vulgaris is the dominant species of an extensive and unique ecosystem of international importance designated as `Calluna heath'. In most parts of western Europe, heathlands are now regressing rapidly because of the cessation of traditional agropastoral practices, which were responsible for their formation, as well as active afforestation, agricultural reclamation and an increase in atmospheric nitrogen deposition.
A previous study on allozyme variation in 18 populations of C. vulgaris from the south-western continental range of the species indicated that these populations were polymorphic at 52.4% of loci, maintained 2.5 alleles per locus and exhibited a mean expected heterozygosity of 0.191 (Mahy et al., 1997). In this part of Calluna's range, a trend of decreasing genetic diversity with distance north was found, which we hypothesized to be related to the postglacial history of the species. Population differentiation was low, with more than 95% of genetic variation found within populations, but a pattern of isolation by distance was detected. Some of the regions examined in this first study included highly fragmented populations (in Belgium and the Spanish Basque country).
The potential influence of distributional shifts during and after the last glacial age is more easily detected by examining populations in territories that were previously glaciated and are currently inhabited by the species of interest (Hewitt, 1996). During the glacial maximum, Scotland was entirely ice-covered, and there is no doubt that C. vulgaris did not survive this period in the region (Godwin, 1975). Today, C. vulgaris may be claimed to be one of the most prevalent and widespread plant species in Scotland. Sixteen per cent of the land is dominated by heather, and 38% (3 × 106 ha) has heather present (Magnusson, 1995).
This study assesses the allozyme variability and genetic differentiation of Scottish C. vulgaris populations in a landscape previously glaciated but currently with a low level of fragmentation compared with continental regions. We compare Scottish populations with previously studied continental populations (Mahy et al., 1997) with the particular aims of examining differences in allozyme variation between the two groups of populations and determining the degree to which Scottish populations are differentiated from continental populations.
Materials and methods
Genetic structure of Scottish populations
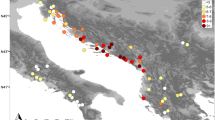
Twelve populations of C. vulgaris were sampled across Scotland (Fig. 1, Table 1). Populations were representative of the range of ecological conditions encountered by the species at low and mid-altitude, of a range of management practices (burned, grazed and unmanaged) and levels of plant vigour (vigorous, grazing suppressed, climatically suppressed). The population at Culbin Forest (Sc9) was mainly composed of the hairy form of C. vulgaris (C. vulgaris var. hirsuta). Thirty-one to 48 individuals were sampled per population, giving a total of 475 individuals.
Location of the 12 Scottish populations of Calluna vulgaris sampled for the study of allozyme diversity and genetic structure, and location of the 18 C. vulgaris populations sampled for the allozyme diversity and genetic structure study in south-western continental regions (Mahy et al., 1997). Sc, Scotland; Be, Belgium; MC, Massif Central (France); La, Landes (France); Py, the Pyrenees; Sp, the Spanish Basque country.
Sampling methods, enzyme extraction and techniques for starch gel electrophoresis were carried out according to the method of Mahy et al. (1997). Seven loci, already studied in the south-western continental populations of C. vulgaris, were examined: Mdh-2 and Mdh-3 (EC 1.1.1.37), Pgm-3 (EC 2.7.5.1), 6pgd-2 (EC 1.1.1.44), Pgi-2 (EC 5.3.1.9), Mnr-1 (EC 2.7.5.1) and Idh-2 (EC 1.1.1.42). The genetic basis of polymorphic patterns was inferred from known subunit composition and the number of isozymes commonly observed in diploid plants. All the variants detected in Scottish populations were compared with the variants reported by Mahy et al. (1997) by running them side-by-side.
Three multilocus measures of genetic diversity were computed: average number of alleles per locus (A); percentage of polymorphic loci at the 0.05 criterion (PLP); and mean expected heterozygosity under Hardy–Weinberg equilibrium (He, gene diversity; Nei, 1978) using GENSURVEY (Vekemans & Lefèbvre, 1997). Deviation from Hardy–Weinberg equilibrium for each variable locus in each population as well as homogeneity of allele frequencies among populations were assessed with unbiased estimates of the Fisher's exact test using GENEPOP (Raymond & Rousset, 1995). Furthermore, to assess the level of differentiation among populations, the global deficit of heterozygotes (FIT) was partitioned into its two components: the deficit of heterozygotes within populations (FIS) and the deficit of heterozygotes among populations (FST) (Wright, 1951). The F-statistics and their 95% and 99% confidence intervals (CI) were estimated using F-STAT (Goudet, 1995) according to the method of Weir & Cockerham (1984). Gene flow between populations was estimated according to the equation: Nm = (1–FST)/4FST (Wright, 1951).
Cavalli-Sforza & Edwards (1967) arc distance was used as the basis for an UPGMA cluster analysis using BIOSYS-1 (Swofford & Selander, 1981). This distance was chosen to follow the method used by Mahy et al. (1997) and because the resulting dendrogram had the highest cophenetic correlation with the data when compared with other classical genetic distances. Geographical and genetic data were also combined in order to test for isolation by distance among the Scottish populations. The extent of gene flow between two populations was estimated for each pair of populations as M= (1–q)/4q, with q the coancestry coefficient (Reynolds et al., 1983; Slatkin, 1993). A log10–log10 linear regression of M against geographical distances for each pair of populations was computed (Slatkin, 1993). Significance of the correlation between the matrix of M and geographical distance after log transformation was tested using a Mantel test with 1000 permutations using GENSURVEY.
Scottish vs. continental populations
To compare Scottish and continental populations, we include the 18 populations (593 individuals) studied by Mahy et al. (1997) in south-western Europe (Fig. 1). Student's t-tests were used to test for significant differences in genetic diversity between Scottish and the entire set of continental populations. Percentage data were arcsine transformed before statistical analysis. In addition, populations were grouped into six regions according to their geographical proximity — Scotland, Belgium, the Massif Central (France), Landes (France), the Pyrenees (France–Spain) and the Spanish Basque country (hereafter designated as Spanish populations) — and averaged measures of genetic diversity (A, PLP, He) were calculated for each region. Genetic relationships among all populations were examined by computing the Cavalli-Sforza & Edwards (1967) arc distance between all pairs of populations and using the genetic distance matrix in an UPGMA cophenetic clustering (BIOSYS-1; Swofford & Selander, 1981). Furthermore, to test whether specific groups of populations were significantly differentiated, we tested the significance of the difference between the mean Reynolds distance (Reynolds et al., 1983) within groups of populations against mean Reynolds distances between groups of populations with the following numerical resampling method (Sokal & Rohlf, 1995) using GENSURVEY: 1000 samples were simulated randomly, assigning populations to each group in order to build the distribution of the statistic under interest, i.e. the difference between average pairwise distance within a group and the average pairwise distance between groups. The observed difference was then tested against two-tailed critical values at 95% and 99% of the distribution.
Results
From the seven loci surveyed, all except Mdh-3 showed some variation in at least one Scottish population. A total of 23 putative alleles were detected across Scottish populations. Two rare alleles detected in Scottish populations were not detected in south-western continental populations: Idh-2.8 coding for a form intermediate between Idh-2.1 and Idh-2.2; and Pgi-2.8 coding for a form intermediate between Pgi-2.1 and Pgi-2.2.
The Scottish within-population genetic variability estimates are presented in Table 1 for each population. Mean proportion of polymorphic loci (PLP) was 40.5 ± 8.2 (SD). The mean number of alleles per locus (A) ranged from 1.6 to 2.3 with a mean of 1.9 ± 0.2. The mean gene diversity (He) varied from 0.087 to 0.167 with a mean of 0.133 ± 0.025. Scottish populations had significantly less mean allozyme diversity than south-western continental populations as a whole (Table 1). Also, Scottish populations appeared to be less variable than all individual continental regions examined for all estimates of genetic diversity (Table 1).
The genotype distribution at each locus in each Scottish population departed significantly from the Hardy–Weinberg equilibrium in only one out of 49 cases, a frequency expected by chance alone. The FIS-values for individual loci were always low, ranging from −0.052 (Idh-2) to 0.059 (Mnr-1), with a mean of 0.020 that was not significantly different from zero (95% CI −0.050 to 0.059).
Significant (P< 0.05) heterogeneity in allelic frequencies among the 12 populations was observed at all variable loci except Mdh-2 (P= 0.360). However, the FST-values for individual loci were low, ranging from −0.001 (Mdh-2) to 0.044 (Pgm-3), with a mean of 0.024, significantly different from zero (99% CI 0.018–0.042). The number of migrants exchanged per generation among the 12 populations (Nm) was estimated as 10.16. The inclusion of European populations in the UPGMA dendrogram based on Cavalli-Sforza & Edwards distances did not modify the relative positions among Scottish populations. Thus, only the dendrogram including all populations (Scottish + continental) is represented in Fig. 2. No clear evidence of geographical ordering was detected among Scottish populations. Adjacent populations did not tend to cluster more frequently than distant populations. The matrix of gene flow (M) and the matrix of geographical distance were not significantly correlated (standardized Mantel statistic, r= 0.066, P= 0.62). This suggested that gene flow between Scottish populations did not decrease with distance separating those populations.
UPGMA cluster analysis of Cavalli-Sforza & Edwards' pairwise distances for Scottish and south-western continental populations of Calluna vulgaris. Geographical location of the populations is shown in Fig. 1.
The UPGMA clustering based on Cavalli-Sforza & Edwards distance suggested that Scottish C. vulgaris populations were reasonably separated from other continental populations, albeit with an individual Belgian population intermingled (Fig. 2). This was consistent with the fact that the mean Reynolds distance among Scottish populations and all other groups of populations (continental and individual continental regions) was significantly higher than the mean pairwise Reynolds distances within groups (Table 2). Despite the fact that Scottish populations formed a homogeneous genetic group, the genetic structure at the scale considered was not shaped by a major discontinuity among Scotland and the continent. Indeed, the UPGMA dendrogram (Fig. 2) exhibited three major sub-branches: one grouping the Scottish and Belgian populations; another the Massif Central and Landes populations; and the third the Pyrenean and Spanish populations. Examination of mean Reynolds distances among specific groups of populations (Table 2) also indicated that the mean genetic distance between Scottish and Belgian populations (0.0354) was far lower than the mean genetic distance between Belgian and either Spanish + Pyrenean populations (0.0594) or Massif Central + Landes populations (0.0591). Altogether, these results suggest that Belgian populations are genetically more similar to Scottish populations than to other continental populations.
Discussion
The partitioning of total genetic diversity into within- and among-population components for Scottish C. vulgaris populations is similar to the pattern reported for continental populations by Mahy et al. (1997). Most of the variation (>95%) is found within populations. Nevertheless, the differentiation among Scottish populations was exceptionally low (FST = 0.024), despite some populations being separated by more than 300 km. The very low differentiation between any of the Scottish populations is confirmed by the lack of consistently interpretable clustering of populations and by the lack of isolation by distance. The estimate of Nm among Scottish populations of C. vulgaris was consistently higher than the threshold value of 1.0 considered sufficient to prevent significant divergence caused by genetic drift for neutral loci (Wright, 1951).
Mahy et al. (1997) suggested that outcrossing, insect pollination and evidence for occasional long-distance dispersal of seeds by wind or animals may reasonably account for high estimates of gene flow between Calluna populations situated within the same geographical region. Some features of the Scottish environment are also likely to enhance the level of gene flow experienced by C. vulgaris populations in this region. Long-distance dispersal of viable seeds in the droppings of red grouse Lagopus lagopus scoticus may be expected to play a significant role in Scotland, where many heaths are managed for red grouse shooting (Welch, 1985). Long-distance dispersal of seed-retaining flowers over smooth, frozen surfaces during the winter can be shown to be possible even in moderate wind conditions (Legg et al., 1992). In general, dispersal of seeds and pollen by wind may be expected to be favoured in the treeless windy environment in which most Scottish C. vulgaris populations occur. Alternatively, if gene movement in this species through pollen or seeds is actually more restricted and does not occur over long distances, then the lack of apparent isolation by distance within the Scottish region may suggest that the species has not reached an equilibrium between genetic drift and gene flow in this part of its range (Slatkin, 1993). Despite the fact that C. vulgaris recolonized Scotland very early in the Holocene (Godwin, 1975), the extension of Calluna heath took place mainly under human activity in historical times (Pennington, 1969), and the number of generations since the establishment of most of the populations may have been too low for genetic drift and differentiation to occur.
In contrast to allozyme markers, which did not reveal much differentiation among Scottish populations of C. vulgaris, ecotypic differences for characteristics of adaptive significance have been observed for growth on toxic soil, for growth form and for floral phenology in Scotland and other areas of Britain (Grant & Hunter, 1962; Bannister, 1978; Marrs & Bannister, 1978). Allozyme and morphological or physiological patterns are not consistent. This suggests that allozymes are mainly selectively neutral genetic markers reflecting the effects of history and chance, whereas patterns of variation for many morphological and physiological characters reflect the effect of natural selection (Ennos, 1995). The genetic structure of the Culbin Forest population (Sc9) gives some support to this conclusion. This population is morphologically well differentiated from other sampled populations because the hairy form of C. vulgaris (C. vulgaris var. hirsuta), usually rare in nature, predominates (Grant & Hunter, 1962). This presumably reflects the effect of selection in an environment with low water potential. However, the allozyme data did not exhibit any significant differentiation.
The low level of allozyme variation observed within Scottish populations of C. vulgaris compared with continental populations seems surprising, as Scotland presently supports the largest, mainly nonfragmented, populations in western Europe, and these populations apparently exchange genes, a feature expected to counteract genetic drift. Furthermore, agreement with Hardy–Weinberg expectations and nonsignificant mean FIS values indicate that inbreeding is not currently an evolutionarily significant process in Scottish populations of C. vulgaris. The effect of late-Quaternary climatological events may provide the most reasonable explanation for the low genetic diversity of Scottish populations and their genetic similarity to Belgian populations. On the continent, populations from regions where C. vulgaris probably did not survive the last glaciation, such as Belgium, also displayed less allozyme variation than populations from presumed refugia. The observation of low allozyme variation within regions where palynological or geological data indicate no survival of C. vulgaris throughout the glacial stage, such as Scotland and presumably Belgium, is consistent with theoretical predictions that founder events and bottlenecks caused during recolonization processes should lead to a loss of genetic diversity (Nei et al., 1975; Hewitt, 1996). The observation that both allelic diversity and heterozygosity are strongly decreased in Scottish populations suggests that the species has gone through a strong historical bottleneck in this region (Nei et al., 1975). The genetic similarity of the Scottish and Belgian populations compared with other continental populations indicates, at least partially, a common evolutionary history for these two sets of populations. This observation is not consistent with an absence of any genetic contact between British and continental populations since the beginning of the last glacial stage, but it is consistent with the proposal of Huntley & Birks (1983) that northern and central parts of the continental species area have been recolonized along a west–east direction from British populations at a period when the British Isles were still a part of continental Europe.
The apparent vigour of C. vulgaris and its extension throughout various environments in Scotland, despite the low allozyme diversity observed in this region, suggests that allozyme variation is not a good indicator of short-term ecological success in this species and that demographic data should be more useful for monitoring and managing populations in the short term.
References
Bannister, P. (1978). Flowering and shoot extension in heath plants of different geographical origin. J Ecol, 66: 117–131.
Cavalli-Sforza, L. L. and Edwards, A. W. F. (1967). Phylogenetic analysis: models and estimation procedures. Evolution, 21: 550–570.
de Beaulieu, J. C. and Reille, M. (1992). Long Pleistocene pollen sequence from the Velay Plateau (Massif-Central, France) I. Ribains Maar. Veget Hist Archaeobot, 1: 233–242.
Ennos, R. A. (1995). Utilising genetic information in plant conservation programmes. In: Hochberg, M. E., Clobert, J. and Barbault, R. (eds) The Genesis and Maintenance of Biological Diversity, pp. 278–291. Oxford University Press, Oxford.
Godwin, H. (1975). History of the British Flora. Cambridge University Press, Cambridge.
Goudet, J. (1995). F-STATv-1.2: a computer program to calculate F-statistics. J Hered, 86: 485–486.
Grant, S. A. and Hunter, R. F. (1962). Ecotypic differentiation of Calluna vulgaris (L.) in relation to altitude. New Phytol, 61: 44–55.
Hamrick, J. L. and Godt, M. J. (1989). Allozyme diversity in plant species. In: Brown, A. H. D., Clegg, M. T., Kahler, A. L. and Weir, B. S. (eds) Plant Population Genetics, Breeding and Genetic Resources, pp. 43–63. Sinauer Associates, Sunderland, MA.
Hewitt, G. M. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc, 58: 247–276.
Huntley, B. and Birks, H. J. B. (1983). An Atlas of Past and Present Pollen Maps for Europe: 0–13000 Years Ago. Cambridge University Press, Cambridge.
Jalut, G., Delibrias, G., Dagnac, J., Mardones, M. and Bouhours, M. (1982). A palaecological approach to the last 21000 years in the Pyrenees: the peat bog of Freychinede (alt 1350m, Ariège, South France). Palaeogeog-Climatol-Ecol, 40: 321–359.
Legg, C. J., Maltby, E. and Proctor, M. C. F. (1992). The ecology of severe moorland fire on the North Yorks Moors: seed distribution and seedling establishment of Calluna vulgaris. J Ecol, 80: 737–752.
Magnusson, M. (1995). Foreword. In: Thompson, D. B. A., Hester, A. J. and Usher, M. B. (eds) Heaths and Moorland: Cultural Landscapes, pp. 135–139. HMSO, Edinburgh.
Mahy, G., Vekemans, X., Jacquemart, A. L. and de Sloover, J. R. (1997). Allozyme diversity and genetic structure in south-western populations of heather, Calluna vulgaris (L.) Hull. New Phytol, 137: 325–334.
Mardonnes, M. and Jalut, G. (1983). La tourbière de Biscaye (alt 409m, Hautes-Pyrénées): approches paléoécologiques des 45000 dernières années. Pollen Spores, 25: 163–212.
Marrs, R. H. and Bannister, P. (1978). The adaptation of Calluna vulgaris (L.) Hull to contrasting soil types. New Phytol, 81: 753–762.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89: 583–590.
Nei, M., Maruyama, T. and Chakaborty, R. (1975). The bottleneck effect and genetic variability in populations. Evolution, 29: 1–10.
Pennington, W. (1969). The History of British Vegetation. English Universities Press, London.
Raymond, M. and Rousset, F. (1995). GENEPOP (version 1.2), a population genetics software for exact tests and ecumenicism. J Hered, 86: 248–249.
Reille, M. and de Beaulieu, J. C. (1988). History of the Würm and Holocene vegetation in Western Velay (Massif-Central, France), a comparison of pollen analysis from three corings at Lac du Bouchet. Rev Palaeobot Palynol, 54: 233–248.
Reynolds, J., Weir, B. S. and Cockerham, C. (1983). Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics, 105: 767–776.
Slatkin, M. (1993). Isolation by distance in equilibrium and non-equilibrium populations. Evolution, 47: 264–279.
Sokal, R. R. and Rohlf, F. J. (1995). Biometry, 3rd edn. W. H. Freeman, New York.
Swofford, D. L. and Selander, R. B. (1981). BIOSYS-1: a Fortran program for the comprehensive analysis of data in population genetics and systematics. J Hered, 72: 281–283.
Vekemans, X. and Lefèbvre, C. (1997). On the evolution of heavy metal tolerant populations in Armeria maritima: evidence from allozyme variation and reproductive barriers. J Evol Biol, 10: 175–191.
Weir, B. S. and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution, 38: 1358–1370.
Welch, D. (1985). Studies in the grazing of heather moorland in north-east Scotland. IV. Seed dispersal and plant establishment in dung. J Appl Ecol, 22: 461–472.
Wright, S. (1951). Evolution in Mendelian populations. Genetics, 16: 97–159.
Young, A., Boyle, T. and Brown, T. (1996). The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol, 11: 413–418.
Acknowledgements
We thank C. Legg for his help in choosing Scottish populations and his comments about the ecology and history of Calluna populations. We thank J. R. De Sloover, X. Vekemans and J. D. Thompson for critical discussions and suggestions on an early version of the manuscript. The study was supported financially by a FDS grant from the Université catholique de Louvain (no. 629108). Sampling in Scotland was supported by a grant from the National Fund for Scientific Research, Belgium, to G.M. A.-L.J. is presently research associate of the National Fund for Scientific Research, Belgium.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mahy, G., Ennos, R. & Jacquemart, AL. Allozyme variation and genetic structure of Calluna vulgaris (heather) populations in Scotland: the effect of postglacial recolonization. Heredity 82, 654–660 (1999). https://doi.org/10.1046/j.1365-2540.1999.00506.x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.1999.00506.x
Keywords
This article is cited by
-
Genetic Diversity and Spatial Genetic Structure of Chamaedaphne calyculata (Ericaceae) at the Western Periphery in Relation to its Main Continuous Range in Eurasia
Folia Geobotanica (2014)
-
Implementation of a model for identifying Essentially Derived Varieties in vegetatively propagated Calluna vulgaris varieties
BMC Genetics (2008)