Abstract
The species flocks of cichlid fish in the three great East African Lakes, Victoria, Malawi, and Tanganyika, have arisen in each lake by explosive adaptive radiation. Various questions concerning their phylogeny have not yet been answered. In particular, the identity of the ancestral founder species and the monophyletic origin of the haplochromine cichlids from the East African lakes have not been established conclusively. In the present study, we used the anonymous nuclear DNA marker DXTU1 as a step towards answering these questions. A 280 bp-fragment of the DXTU1 locus was amplified by the polymerase chain reaction from East African lacustrine species, the East African riverine cichlid species Haplochromis bloyeti, H. burtoni and H. sparsidens, and other African cichlids. Sequencing revealed several indels and substitutions that were used as cladistically informative markers to support a phylogenetic tree constructed by the neighbor-joining method. The topology, although not supported by high bootstrap values, corresponds well to the geographical distribution and previous classification of the cichlids. Markers could be defined that: (i) differentiate East African from West African cichlids; (ii) distinguish the riverine and Lake Victoria/Malawi haplochromines from Lake Tanganyika cichlids; and (iii) indicate the existence of a monophyletic Lake Victoria cichlid superflock which includes haplochromines from satellite lakes and East African rivers. In order to resolve further the relationship of East African riverine and lacustrine species, mtDNA cytochrome b and control region segments were sequenced. The mtDNA-based trees support the notion of the monophyly of the Lake Victoria superflock but are ambigious with respect to the phylogenetic position of the Lake Malawi flock.
Similar content being viewed by others
Introduction
The three great lakes in the East African Rift Valley, Lakes Victoria, Tanganyika, and Malawi, each harbour a species flock of cichlid fish (family Cichlidae, suborder Labroidei, order Perciformes, Kaufman & Liem, 1982). Each flock is comprised of several hundred species (Fryer & Iles, 1972). Each is believed to be monophyletic (Kornfield, 1978; Meyer et al., 1990; Greenwood, 1991; Meyer, 1993), and all three flocks have been claimed to be derived from a single common ancestor (Kornfield, 1991). The flocks are of different ages, but all seem to have arisen in situ in each lake after its formation or re-formation (Fryer & Iles, 1972). The age estimates for the three lakes are 12 million years (Myr) for Lake Tanganyika (Cohen et al 1993), 2 Myr for Lake Malawi (Fryer & Iles, 1972) and 12 400 years for Lake Victoria (Johnson et al., 1996). The speed with which adaptive radiation produced the flocks is therefore truly amazing, especially in Lake Victoria. At the same time, the diversity of forms constituting the flocks is astounding. In each lake, the fish fall into several trophic groups specialized for very different food resources (Fryer & Iles, 1972), and within each group the species differ in body size, body shape, the morphology of their body parts (in particular the jaw apparatus, colouration and behaviour; (Poll, 1986; Greenwood, 1991; Ribbink, 1991; Sturmbauer & Meyer, 1992).
Although the East African cichlids have received a great deal of attention from evolutionary biologists, several questions regarding their evolution remain unanswered. Prime among these are still the questions of the monophyly of the species flocks in the great lakes and the origins of the ancestral species that founded them. Lake Tanganyika is the oldest of the three lakes (Cohen et al., 1993) and its cichlids are classified into 12 tribes (Poll, 1986; Nishida, 1991; Sturmbauer & Meyer, 1992). The ancestors of the species flocks of the younger Lakes Malawi and Victoria are believed to have reached the lakes through the riverine drainage systems. But which species were the founder species and are the species flocks really monophyletic? The present study has been aimed at providing an answer to this question using cichlid samples available to us.
Materials and methods
Fish
Cichlid fish were collected in East Africa during expeditions in October/November 1993 and in October 1995. Additional samples were purchased from dealers (Aquaristik Pelz, Bondorf, Germany, and Kölle Zoo, Stuttgart, Germany) or were kindly provided by Dr Axel Meyer (Department of Ecology and Evolution, State University of New York, Stony Brook, USA) and Dr Lothar Seegers (Dinslaken, Germany). The species and their origins are listed in Table 1.
Preparation of genomic DNA
Total genomic DNA was isolated from hepatopancreases fixed in 70% ethanol after excision from freshly caught fish. The isolation was carried out using the QIAamp tissue Kit (QIAGEN, Hilden, Germany). Contaminating RNA was removed by digestion with RNase A (30 min at 37°C), followed by phenol/chloroform extraction. The concentration of the DNA was adjusted to 100 μg mL−1.
Oligonucleotides
The oligonucleotide primers for DXTU1 (R3: 5′-ACGGTGTGCGTCTCTATATCTGGTG-3′ and R4: 5′-GCCATTTCGAGCAAGGTTAATG-3′), for the mitochondrial (mt) DNA control region (L15926: 5′-TCAAAGCTTACACCAGTCTTGTAAACC-3′ and H16498: 5′-CCTGAAGTAGGAACCAGATG-3′), and for the mtDNA cytochrome b gene (L14724: 5′-CGAAGCTTGATATGAAAAACCATCGTTG-3′ and H15149: 5′-AAACTGCAGCCCCTCAGAATGATATTTGTCCTCA-3′, Kocher et al., 1989; Meyer et al., 1990) were synthesized by Pharmacia (Freiburg, Germany).
Polymerase chain reaction (PCR)
The reaction (Saiki et al., 1985, 1988) was carried out in the PTC-100 Programmmable Thermal Controller (MJ Research, Watertown, MA, USA, and Biozym, Hess. Oldendorf, Germany) in a volume of 25 μL containing 1×reaction buffer (Pharmacia, Freiburg, Germany), 100 μM each of dATP, dCTP, dGTP and dTTP, 1 μM of each of the appropriate primers, 1.25 U of Taq polymerase (Pharmacia) and 50 ng of total genomic DNA. The programme consisted of 3 min initial denaturation at 94°C, followed by 35 cycles with 15–30 s denaturation at 94°C, 15–30 s primer annealing at 52°C–55°C, and 1–2 min primer extension at 72°C. The reaction was completed by a primer extension step for 5 min at 72°C. For DNA that did not amplify under these conditions, reaction parameters were optimized using the HotWax OptiStart Kit (Invitrogen, Leek, The Netherlands). Subsequent reactions were performed in 1×PCR buffer, pH 9.0 (primers R3 and R4) or pH 8.5 (primer pairs L14724/H15149 and L15926/H16498), 2.5 mM HotWax Mg2+ Beads (Invitrogen), 500 μM each of dATP, dCTP, dGTP, and dTTP, 1 μM of each of the primers, 2.5 U of Taq polymerase (Pharmacia), and 50 ng of total genomic DNA.
Subcloning of PCR products
The PCR products were separated by electrophoresis in 1% low melting point agarose (Gibco BRL, Gaithersburg, MD, USA). Gel slices containing the expected fragments were excised, the DNA was eluted by centrifugation in a microfuge, and ligated into the SmaI site of the vector pUC18 using the SureClone ligation kit (Pharmacia). Competent E. coli XL1-Blue (Stratagene) were used for transfection. Alternatively, the PCR products were extracted from gel slices with the QIAquick Gel Extraction Kit (QIAGEN) and cloned using the pT7Blue T-Vector Kit (Novagen and AGS, Heidelberg, Germany).
Sequencing of PCR products
The DNA was sequenced by the dideoxy chain-termination method (Sanger et al., 1977) using the AutoRead sequencing kit (Pharmacia) with vector-derived fluorescent primers and the A.L.F. DNA Sequencer (Pharmacia).
Sequence analysis
The sequences were analysed using the SEQSPEAK 1.0 DNA Sequence Editor for Macintosh (written by Keith Conover, Dalhousie University, Halifax, Canada 1991) and aligned using the SEQPUP 0.6e program (written by Don G. Gilbert, Indiana University, Bloomington, IN, USA 1996).
Construction of dendrograms
Genetic distances were calculated by Kimura's two-parameter method (Kimura, 1980), and phylogenetic reconstruction using the neighbor-joining method (Saitou & Nei, 1987) was performed by the MEGAsoftware, Version 1.0 (Kumar et al., 1993) with the pairwise deletion option for gaps. The reliability of the tree topology was assessed by 500 bootstrap replications (Felsenstein, 1985).
Results
Variability at the DXTU1 locus
DXTU1 is an anonymous nuclear DNA marker originally identified by the random amplification of polymorphic DNA (RAPD) technique (Welsh & McClelland, 1990; Williams et al., 1990) and shown to vary among the East African cichlid species (Sültmann et al., 1995). Variant-specific PCR experiments indicated the presence of only a single locus (Sültmann et al., 1995). To PCR amplify the most variable part of the DXTU1 locus, we used the primer pair R3/R4 and genomic DNAs of East, Central, and West African, as well as South American cichlid species as templates. The 280 bp-long amplified fragments were then cloned and sequenced. To minimize sequencing errors, at least three independent clones of each amplified fragment were sequenced. Altogether, we sequenced DXTU1 fragments from 46 fishes representing 25 species. The alignment of 44 sequences, together with selected sequences from Sültmann et al. (1995), is shown in Fig. 1.
Alignment of representative DXTU1 nucleotide sequences. The sequences obtained in this study with the primer pair R3 and R4 are shown together with sequences described previously by Sültmann et al. (1995; indicated by a ‘•’ symbol). Dashes indicate identity with the consensus sequence, asterisks indicate deletions, and dots missing sequence information. Designations are explained in Table 1. The positions of informative indels (ID) and substitutions (SB) are indicated by arrows. The nucleotide sequences reported in this paper have been submitted to the GenBank database and have been assigned the accession numbers AF014961-AF015004.
The sequences were evaluated by two methods. First, the sequence alignment was used to construct a phylogenetic tree by the neighbor-joining method (Saitou & Nei, 1987) using the pairwise deletion option for insertions/deletions (indels) to obtain the genetic distances; and secondly, the indels were used as cladistic markers and added onto the tree obtained by the first method (Fig. 2). Viewed without the indel markers, the phylogenetic tree is not very robust. Although some branches are clearly differentiated by high bootstrap values, others are differentiated poorly by low bootstrap values. The addition of the indel markers onto the tree, however, considerably enhances its robustness and by delineating certain groups, increases the confidence in the overall topology of the tree. Seven cladistically informative markers were identified and numbered arbitrarily from 1 to 7. Their positions and lengths are apparent from Fig. 1. The sequence alignment also reveals several diagnostic nucleotide substitutions. These, too, are numbered, and their distribution is shown in the tree in Fig. 2. (In contrast to the indels, the diagnostic substitutions were included in the data from which the tree was constructed.) Together, the indels and the diagnostic substitutions form a nested hierarchy which corresponds well to the geographical distribution of the various taxa and/or to previous classification of the cichlid fishes based on other types of data (Meyer et al., 1990; Stiassny, 1991; Sturmbauer & Meyer, 1992; Meyer, 1993).
Neighbor-joining tree based on the DXTU1 locus sequences. Genetic distances were calculated by Kimura's two-parameter method (Kimura, 1980). The numbers at each node represent the percentage recovery of the node in 500 bootstrap replications. The origin of the species and the clades defined by informative indels and substitutions (see text) are indicated
In the tree, the Lake Victoria and satellite lake haplochromines are divided into two clusters, one encompassing the species Haplochromis pyrrhocephalus, H. plagiodon, H. sauvagei, H. venator and H. velifer, and the other the species H. nigricans, H. sauvagei and H. brownae. The latter cluster also includes two riverine species, H. sparsidens and H. ‘chala’. However, the two clusters are differentiated by single nucleotide substitutions only and hence supported by low bootstrap values. The riverine species H. bloyeti and Schwetzochromis malagaraziensis form an outgroup to the entire Lake Victoria flock plus H. sparsidens. This clade 1 is supported by the diagnostic substitution SB5 (Fig. 2). The Lake Malawi species, together with two riverine species (Ctenochromis pectoralis, H. burtoni) and one Lake Tanganyika species (Cyprichromis ‘mpulungu’), are united with clade 1 into clade 2 by indel ID1. All H. burtoni individuals share the diagnostic substitution SB6. The remaining Lake Tanganyika species are united with clade 2 into clade 3 by indel ID2 and the diagnostic substitution SB1. Lake Tanganyika species (with the exception of C. ‘mpulungu’), the West African riverine species Steatocranus casuarius, as well as the species of the genera Tilapia and Oreochromis, are united in group 4 by the diagnostic substitution SB4. The tilapiines (with the exception of T. sparrmanni), oreochromines, Central and South American species, and the West African species Pelvicachromis humilis, P. pulcher, Tylochromis leonensis, Hemichromis fasciatus, H. bimaculatus and H. lifalili are united by the diagnostic substitution SB3. The same group, with the exclusion of the tilapias, is also characterized by the diagnostic substitution SB2. The Pelvicachromis species share indels ID6 and ID7; the Hemichromis species share indels ID4 and ID5; and Hemichromis lifalili and H. bimaculatus share indel ID3.
Variability of mtDNA
Additional support for the relationships among the cichlid taxa as seen from the DXTU1 data, was sought in mtDNA sequences. MtDNA has been used by other investigators (Meyer et al., 1990; Sturmbauer & Meyer, 1992, 1993) to determine phylogenetic relationships among the cichlid species of the East African great lakes; the river species and the species from the smaller lakes have been studied much less extensively. We amplified the mtDNA cytochrome b and a control region segment by using the primer pairs L14724 — H15149 and L15926 — H16498, respectively. The PCR with the former pair yielded a 420 bp fragment which encompassed most of the cytochrome b gene; using the latter pair, a 490 bp fragment was obtained which encompassed part of the tRNA-Thr gene, the tRNA-Pro gene and the first hypervariable segment of the control region. The fragments were cloned and sequenced and the sequences were used to produce phylogenetic trees (Figs. 3 and Fig. 4).
Neighbor-joining tree based on representative mitochondrial DNA cytochrome b gene sequences. The sequences obtained in this study were from H. bloyeti, H. ‘chala’, H. sparsidens, H. burtoni, H. velifer, H. brownae, C. pectoralis, S. malagaraziensis, T. rendalli, S. casuarius, O. malagarasi, H. bimaculatus, P. humilis, P. pulcher and T. leonensis. Other sequences were taken from Meyer et al., 1990 or Sturmbauer & Meyer, 1993 or were retrieved from the GenBank database (indicated by locus name); they are as follows: Bathybates ferox (Bathibatini), MIBFCYTBA; Grammatotria lemarii (Ectodini), MIGLCYTBA; Limnochromis auritus (Limnochromini), MILACYTBA; Oreochromis tanganicae (Tilapiini), MIOCYTB; Paracyprichromis brieni (Cyprichromini), MIPBCYTB; Perissodus straeleni (Perissodini), MIPSCYTB; Tanganicodus irsacae (Eretmodini), MITICYTBB; Tropheus duboisi, (Tropheini), MITDCYTBJ; Tropheus moorii (Tropheini), MITMCYTBB. ‘Haplochromine A’ and ‘Haplochromine B’ are cytochrome b sequences from Lake Malawi sand dweller and rock dweller cichlids, respectively; ‘Haplochromine C’ is from Lake Victoria, (Meyer et al., 1990). The species abbreviations are explained in Table 1. The numbers at each node represent the percentage recovery of the node in 500 bootstrap replications. The nucleotide sequences reported in this paper have been submitted to the GenBank database and have been assigned the accession numbers AF015017-AF015036.
Neighbor-joining tree based on representative mitochondrial DNA control region sequences. The sequences obtained in this study were from H. bloyeti, H. ‘chala’, H. sparsidens, H. burtoni, H. velifer, O. malagarasi and P. humilis. Published sequences. (Meyer et al., 1990; Sturmbauer & Meyer, 1993) were retrieved from the GenBank database (indicated by locus name); they are as follows: Bathybates ferox (Bathibatini), MIBFTRNGX; Grammatotria lemarii (Ectodini), MIGLTRNGX; Limnochromis auritus (Limnochromini), MILATRNGX; Oreochromis tanganiae (Tilapiini), MIOTTRNGX; Paracyprichromis brieni (Cyprichromini), MIPBTRNGX; Perissodus straeleni (Perissodini), MIPSTRNGX; Tanganicodus irsacae (Eretmodini), MITITRNGX; Tropheus moorii (Tropheini), MITMTTPAA; Astatotilapia calliptera, MIHCTRN; Haplochromis burtoni, MIABTRNGX. ‘Haplochromine A’ and ‘Haplochromine B’ are control region sequences from Lake Malawi sand dweller and rock dweller cichlids, respectively; ‘Haplochromine C’ is from Lake Victoria (Meyer et al., 1990). The species abbreviations are explained in Table 1. The numbers at each node represent the percentage recovery of the node in 500 bootstrap replications. The nucleotide sequences reported in this paper have been submitted to the GenBank database and have been assigned the accession numbers AF015005-AF015016.
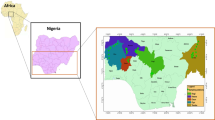
The overall topologies of the two mtDNA trees are concordant with each other and with the tree based on the DXTU1 data (Fig. 2.) The mtDNA-based trees, like the DXTU1-based tree, show a series of nested relationships corresponding to the geographical distribution of the tested species. For an easier comprehension, the geographical areas occupied by the ‘nests’ or groups (Figs. 3 and 4) have been indicated (see also Fig. 5 for a map of East Africa). Thus, group 1 encompasses haplochromines from Lake Victoria and its satellites (e.g. Lake Nabugabo). Group 2 includes group 1 and H. burtoni from the Malagarasi river system, as well as H. sparsidens, H bloyeti and H. ‘chala’ from Lake Manyara, the Wami River system and Lake Chala. Group 3 includes group 2 and Lake Malawi species. Group 4 includes group 3, the widely distributed species Astatoreochromis alluaudi and Ctenochromis pectoralis from the Pangani River, Schwetzochromis malagaraziensis from Malagarasi, as well as Lake Tanganyika species. Group 5 includes group 4, as well as the Oreochromis species. The West African riverine species of the genera Hemichromis, Pelvicachromis and Tylochromis form a set of outgroups to group 5. The nodes of the individual clades are supported by modest-to-high bootstrap values.
Geography of East Africa showing the localities of the cichlid species investigated in this study. The drainage systems occupied by the clades of Figs 3 and 4 are differentiated by shadings. A, Lake Victoria; B, Lake Malawi; C, Lake Tanganyika; D, Lake Manyara; E, Lake Chala; F, Lakes Nabugabo and Kayugi.
Discussion
The combined use of nuclear (DXTU1) and mtDNA (cytochrome b and control region) markers provides a general outline for the evolution of the African cichlids. The DXTU1 data support the notion that ancestral Cichlidae lived in West African lakes and rivers before the separation of the African and South American (sub)continents some 100 Myr ago (Stiassny, 1987). Although the phylogenetic tree in Fig. 2 is unrooted, it seems reasonable to place the root at its base. This placement is supported by other molecular (Meyer, 1993; Zardoya et al., 1996) and morphological (Stiassny, 1991) data, which provide evidence for an outgroup position of Madagascan and Indian cichlids relative to the African and Neotropical species. Indeed, the DXTU1-specific PCR primers, which amplify DNA from African and some South American species (data not shown), fail to amplify DNA from Madagascan species, presumably because the sequence of the latter is too divergent from the former.
The DXTU1 data suggest that the group of West African cichlids, represented by the genera Hemichromis, Pelvicachromis and Tylochromis, diverged first from the common stock, and was then followed by the divergence of the Oreochromis and Tilapia-Steatocranus branches (Fig. 2).
The bootstrap support for the relationships among the main groups of East African taxa is weak. The groups are, however, supported by the sharing of diagnostic indels. Thus, all the tested East African cichlids share indel 2; East African cichlids with the exclusion of those in Lake Tanganyika share indel 1; Lake Victoria and some riverine haplochromines share substitution 5. These markers suggest a single ancestor for the East African cichlids to the exclusion of Lake Tanganyika species and a monophyletic origin of the Lake Victoria superflock (Meyer, 1993) including Haplochromis species from Lake Victoria and its satellite lakes as well as riverine haplochromines.
The mtDNA-based trees identify a group of East African riverine species that are closely related to both the Lake Victoria and Lake Malawi species flocks and are more distantly related to the Lake Tanganyika species (Figs 3 and 4). The group includes H. sparsidens, H. ‘chala’, H. bloyeti and H. burtoni. The trees do not identify clearly any one of these species as being most closely related to the Lake Victoria or the Lake Malawi species and hence as being closest to the putative ancestor that founded the two species flocks. Thus, the cytochrome b-based tree favours H. burtoni as a closest relative of the Lake Victoria species and any of the riverine species as a closest relative of the Lake Malawi species, but the control region-based tree suggests a sister-group relationship of the Lake Malawi species to the riverine and Lake Victoria species (Figs 3 and 4). In the latter instance, the sister-group relationship is supported moderately well by bootstrapping, but in the former instance the bootstrap values of the relevant nodes are low. Moreover, the trees reveal unnatural groupings among the riverine species in which some individuals identified on morphological grounds and by their distribution as H. bloyeti, cluster with H. sparsidens and H. ‘chala’, whereas other individuals, presumably of the same species, form a separate cluster. Both clusters are supported by very high bootstrap values. This result suggests that the populations of some of the riverine species may be genetically, if not morphologically, drifting apart from one another, and further that it is insufficient to analyse only a few or a single individual of the young and rapidly evolving cichlid species. The DXTU1-based tree clusters — again with low bootstrap values — the above riverine species with the Lake Victoria flock, separately from the Lake Malawi flock. The main factor responsible for this clustering is the diagnostic substitution SB5 which is shared by H. bloyeti, H. sparsidens, H. ‘chala’, Schwetzochromis and the Lake Victoria species, but is absent in the Lake Malawi species and in H. burtoni. Additional support for this clustering is provided by an indel discovered recently in the 3′ untranslated region of the glucose-phosphatase gene (S. Nagl, W. Mayer, H. Tichy and J. Klein, in prep.). The indel is present, as a polymorphism, in some of the riverine species in the vicinity of Lake Victoria and all Lake Victoria species thus far tested, but is absent in the Lake Malawi (and Lake Tanganyika) species. These data support the notion that the riverine species H. bloyeti, H. burtoni, H. sparsidens and H. ‘chala’ are more closely related to the Lake Victoria than to the Lake Malawi flocks, although the difference in the degree of relatedness is rather small. The closeness could signify either that the riverine species provided an ancestor or ancestors for the Lake Victoria flock more recently than they did for the Lake Malawi flock or that genes are being exchanged between lacustrine and riverine species (all riverine species tested come from rivers in the geographical area of Lake Victoria). Be this as it may, no single, clearly identifiable candidate for the position of a species ancestral to the Lake Victoria flock has emerged from this group of riverine species, but the data strengthen the previous notion of monophyly of an extended Lake Victoria superflock including satellite lakes and East African riverine cichlids (Meyer, 1993). The relationship to the Lake Malawi flock is not unambiguous. The mitochondrial control region tree suggests a sister-group relationship, but the DXTU1 and cytochrome b data open the possibility that the Lake Malawi flock is part of the superflock. To overcome the uncertainty in the crucial phylogenetic position of the riverine cichlids, clarification by analysing additional markers and larger samples is necessary.
The two remaining riverine species, Ctenochromis pectoralis and Astatoreochromis alluaudi, form separate lineages that seem to have diverged from an ancestral stock before the radiation of the Lake Victoria and Lake Malawi flocks, but after the divergence of the different Lake Tanganyika lineages. Their position in the mtDNA trees is supported by moderately high bootstrap values.
References
Cohen, A. S., Soreghan, M. J. and Scholz, C. A. (1993). Estimating the age of formation of lakes: An example from Lake Tanganyika, East African rift system. Geology, 21: 511–514.
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 39: 783–791.
Fryer, G. and Iles, T. D. (1972). The Cichlid Fishes of the Great Lakes of Africa. TFH Publications, Neptune City, NJ.
Greenwood, P. H. (1991). Speciation. In: Keenleyside, M. H. A. (ed.) Cichlid Fishes. Behaviour, Ecology and Evolution, pp. 86–102. Chapman and Hall, London.
Johnson, T. C., Scholz, C. A., Talbot, M. R., Kelts, K., Ricketts, R. D., Ngobi, G. et al (1996). Late Pleistocene desiccation of Lake Victoria and rapid evolution of cichlid fishes. Science, 273: 1091–1093.
Kaufman, L. and Liem, K. F. (1982). Fishes of the suborder Labroidei (Pisces: Perciformes): phylogeny, ecology and evolutionary significance. Breviora, 472: 1–19.
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol, 16: 111–120.
Kocher, T. D., Thomas, W. K., Meyer, A., Edwards, S. V., Pääbo, S., Villablance, F. X. et al (1989). Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc Natl Acad Sci USA, 86: 6196–6200.
Kornfield, I. L. (1978). Evidence for rapid speciation in African cichlid fishes. Experientia, 34: 335–336.
Kornfield, I. L. (1991). Genetics. In: Keenleyside, M. H. A. (ed.) Cichlid Fishes. Behaviour, Ecology and Evolution, pp. 103–128. Chapman and Hall, London.
Kumar, S., Tamura, K. and Nei, M. (1993). MEGA: Molecular evolutionary genetic analysis. version 1.0. Pennsylvania State University, University Park, PA.
Meyer, A. (1993). Phylogenetic relationships and evolutionary processes in East African cichlid fishes. Trends Ecol Evol, 8: 279–284.
Meyer, A., Kocher, T. D., Basasibwaki, P. and Wilson, A. C. (1990). Monophyletic origin of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature, 347: 550–553.
Nishida, M. (1991). Lake Tanganyika as an evolutionary reservoir of old lineages of East African cichlid fishes: Inferences from allozyme data. Experientia, 47: 974–979.
Poll, M. (1986). Classification des Cichlidae du lac Tanganyika: Tribus, genres et espèces. Mém Acad R Belg Cl Sci, 45: 5–163.
Ribbink, A. J. (1991). Distribution and ecology of the cichlids of the African Great Lakes. In: Keenleyside, M. H. A. (ed.) Cichlid Fishes. Behaviour, Ecology, and Evolution, pp. 36–102. Chapman and Hall, London.
Saiki, R. K., Scharf, S. J., Faloona, F., Mullis, K. B., Horn, G. T., Erlich, H. A. and Arnheim, N. (1985). Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science, 230: 1350–1354.
Saiki, R. K., Gelfand, D. H., Stoffel, S., Scharf, S. J., Higuchi, R., Horn, G. T.et al (1988). Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science, 239: 487–491.
Saitou, N. and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol, 4: 406–425.
Sanger, F., Nicklen, S. and Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA, 74: 5463–5467.
Stiassny, M. L. J. (1987). Cichlid familial interrelationships and the placement of the neotropical genus Cichla (Perciformes, Labroidei). J Nat Hist, 21: 1311–1331.
Stiassny, M. L. J. (1991). Phylogenetic interrelationships of the family Cichlidae: an overview. In: Keenleyside, M. H. A. (ed.) Cichlid Fishes. Behaviour, Ecology and Evolution. pp. 1–35. Chapman and Hall, London.
Sturmbauer, C. and Meyer, A. (1992). Genetic divergence, speciation and morphological stasis in a lineage of African cichlid fishes. Nature, 358: 578–581.
Sturmbauer, C. and Meyer, A. (1993). Mitochondrial phylogeny of the endemic mouthbrooding lineages of cichlid fishes from Lake Tanganyika in Eastern Africa. Mol Biol Evol, 10: 751–768.
Sültmann, H., Mayer, W. E., Figueroa, F., Tichy, H. and Klein, J. (1995). Phylogenetic analysis of cichlid fishes using nuclear DNA markers. Mol Biol Evol, 12: 1033–1047.
Welsh, J. and McClelland, M. (1990). Fingerprinting genomes using PCR with arbitrary primers. Nucl Acids Res, 18: 7213–7218.
Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A. and Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res, 18: 6531–6535.
Zardoya, R., Vollmer, D. M., Craddock, C., Streelman, J. T., Karl, S. and Meyer, A. (1996). Evolutionary conservation of microsatellite flanking regions and their utility in resolving the phylogeny of cichlid fishes (Pisces: Perciformes). Proc R Soc B, 263: 1589–1598.
Acknowledgements
We thank Dr Axel Meyer, Department of Ecology and Evolution, State University of New York, Stony Brook, USA, for DNA samples; Dr Lothar Seegers, Dinslaken, Germany, for providing us with samples of riverine cichlids; Tanja Breitsprecher, Beate Pömmerl and Ljubica Sanader for excellent technical assistance; and Moira Burghoffer for editorial assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mayer, W., Tichy, H. & Klein, J. Phylogeny of African cichlid fishes as revealed by molecular markers. Heredity 80, 702–714 (1998). https://doi.org/10.1046/j.1365-2540.1998.00347.x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.1998.00347.x
Keywords
This article is cited by
-
East African cichlid lineages (Teleostei: Cichlidae) might be older than their ancient host lakes: new divergence estimates for the east African cichlid radiation
BMC Evolutionary Biology (2019)
-
Gene duplications and the evolution of c-type lysozyme during adaptive radiation of East African cichlid fish
Hydrobiologia (2017)
-
Mitochondrial phylogeny and phylogeography of East African squeaker catfishes (Siluriformes: Synodontis)
BMC Evolutionary Biology (2006)
-
Comparative Phylogenetic Analyses of the Adaptive Radiation of Lake Tanganyika Cichlid Fish: Nuclear Sequences Are Less Homoplasious But Also Less Informative Than Mitochondrial DNA
Journal of Molecular Evolution (2005)