Abstract
As a result of human activities natural environments have been altered in many different ways. One important effect of human disturbance is the fragmentation of natural habitats. As a consequence, genetic differentiation among habitat islands is expected to increase, whereas within-area genetic diversity is expected to decrease. Indirect estimates from allozyme polymorphisms are used to investigate the effects of habitat fragmentation in the winter moth on a very small geographical scale. We demonstrated that genetic differentiation increased whereas genetic diversity decreased with fragmentation, with habitat patches isolated by only a few hundred metres up to 3 km. These results were even more striking considering that no genetic differentiation was detected at a larger scale (10–40 km). This pattern of distribution of genetic variation is in agreement with temporarily variable densities and gene flow levels which prevent an equilibrium being reached between genetic drift and gene flow. Consequently the effects of fragmentation probably remain limited.
Similar content being viewed by others
Introduction
Many formerly continuous areas of natural habitat have been subdivided into relatively small so-called habitat islands surrounded by human-altered environments (Skole & Tucker, 1993). As a consequence, many populations have become artificially subdivided into relatively small subpopulations with a limited interchange of individuals. With increasing isolation of subpopulations, interchange of individuals will often decrease and subpopulation dynamics will become increasingly independent. This results in a reduction of the effective population sizes (Ne) and consequently in an increase of the subpopulations' susceptibility to stochastic effects. Three sources of stochastic processes can be distinguished, demographic, environmental and genetic, which may lead to an increased subpopulation extinction risk (Frankham, 1995).
As a result of random genetic drift, genetic diversity decreases within and genetic variation increases among subpopulations (Hartl & Clarke, 1989) as Ne decreases. Temporal reductions in Ne, for example after a local extinction and recolonization event, may also enforce the effects of genetic drift (Wade & McCauley, 1988). Because the reduction of genetic diversity may promote inbreeding depression and may decrease the populations' ability to adapt to a changing environment, it is expected to increase extinction probabilities (Ellstrand & Elam, 1993; Frankham, 1995). Genetic drift is therefore of great interest to conservation biologists. Presumed selectively neutral genetic markers, such as allozymes, restriction fragment length polymorphisms and microsatellites, have been widely used to describe population structure (e.g. Michalakis et al., 1993). This study aims at investigating the effects of habitat fragmentation on the distribution of genetic variation in the winter moth (Operophtera brumata L.).
The winter moth inhabits pedunculate oak (Quercus robur L.) woods which have become highly fragmented in northern Belgium, as in other parts of the world. As adult winter moths show very little dispersal (Graf et al., 1995; Van Dongen et al., 1996; Van Dongen, 1997) this species can be expected to be prone to the adverse effects of fragmentation of its habitat on its genetic population structure. Caterpillars appear to have high dispersal abilities, however, as they can float with the wind for several hundred metres (Edland, 1971), which may counteract the effects of habitat fragmentation.
In a preliminary population genetic study Van Dongen et al. (1994) showed a significant genetic differentiation of winter moth subpopulations on a relatively small geographical scale (<10 km). But this genetic differentiation pattern may have resulted from factors other than fragmentation of the oak woods. With restricted dispersal, genetic differentiation is expected to accumulate over time as a result of the geographical distance among different study sites (i.e. ‘isolation by distance’; Wright, 1943). To investigate if the observed genetic differentiation results from fragmentation of the habitat or merely a distance effect, the genetic population structure should be compared between continuous and fragmented areas. In this paper we investigate the effects of habitat fragmentation on the distribution of genetic variation both within (genetic diversity) and among (genetic differentiation) local subpopulations. We compare genetic diversity and differentiation among collection sites between continuous and fragmented areas on a comparable geographical scale. We relate genetic diversity to the degree of fragmentation of the different woodland fragments.
Materials and methods
Winter moth
The winter moth is a univoltine species whose adults emerge from the soil in November and December. At night the brachypterous females crawl up tree trunks where they chemically attract the winged males for copulation. After copulation males return to the ground whereas females continue climbing the tree trunk and lay their eggs in the canopy. The eggs hatch in early spring. The caterpillars feed upon young leaves for about six weeks, drop to the ground on a silk thread, burrow into the ground, spin a cocoon and pupate (Varley et al., 1973). Pedunculate oak is the primary host on which the winter moth attains the highest feeding success (Wint, 1983), but caterpillars can also feed upon an extensive range of other broad-leaved trees and shrubs, and also on Sitka Spruce (Stoakley, 1995).
Study areas and sampling procedure
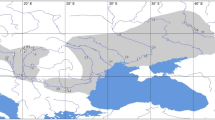
Male winter moths were collected during November 1993 and 1994 in 28 study sites (Table 1, Fig. 1) located in northern Belgium. In some areas males could be collected in one year only, owing to the low moth densities, so that a total of 41 samples was obtained. A total of 1361 males were caught and analysed. Males were either collected by hand between 18.00 and 20.00 hours and/or by means of sticky tree bands as described by Van Dongen et al. (1994). Moths were collected at sites dominated by pedunculate oak.
Map of the 28 study sites for Operophtera brumata in Belgium grouped in five regions (see also Table 1). Ellipses indicate study sites in a continuous landscape.
The 28 sampling sites were grouped in five ‘regions’ (Fig. 1). Those in the regions Boshoek (B), Herentals (H) and Turnhout (T) were fragmented woodlands mainly surrounded by agricultural land and residential areas. Woodlands dominated by other tree species occur relatively sparsely and support low winter moth densities (S. Van Dongen, unpubl. obs.). Surface areas of the woodland fragments varied between 0.5 and 17 ha (Table 1). The Peerdsbos (P) region is a large forest area (>200 ha). In the Schietveld (S) region sampling sites KA and NB were located in a large continuous oak forest (40 ha, Table 1: subregion S1) and ST and SD were two woodland fragments (subregion S2). For the comparison of the degree of genetic differentiation among sampling sites between the continuous and the fragmented environments (see below) the B region was further subdivided into three subregions (B1, B2 and B3) (Table 1) such that the geographical distances between sites within a subregion were comparable to the distances between sites in the continuous forests. Within the regions in the fragmented landscape more than 80 per cent of the oak woodlands were sampled, and most of the unsampled areas support low winter moth densities.
Allozyme electrophoresis
Individual genotypes of the male moths were determined with vertical polyacrylamide gel electrophoresis (PAGE) of five polymorphic enzyme loci: GPI (EC 5.3.1.9), HBDH (1.1.1.30), PEP (3.4.11. * (leucine-alanine)), PGM (5.4.2.2) and PGD (1.1.1.43). Enzyme stainings were adopted from Harris & Hopkinson (1976). Sample preparations and electrophoretic runs were performed as described by Van Dongen et al. (1994).
Genetic population structure
Isolation by distance
Slatkin (1993) showed that isolation by distance can be tested by investigating the relationship between pairwise gene flow estimates and geographical distance on a logarithmic scale. Furthermore, in some cases nonequilibrium patterns may be detected (Slatkin, 1993). Under the isolation by distance model, a linear negative relationship between the logarithm of the pairwise gene flow (Nem) estimates and the logarithm of the geographical distance is expected. However, when genetic differentiation among sites is relatively low a number of pairwise Nem estimates cannot be estimated because the corresponding pairwise value of θ estimates may be negative (Weir, 1996). Therefore, we used θ (as an estimator of FST, Weir, 1996) instead of Nem to test isolation by distance for which a positive relationship is expected (Slatkin, 1993). Because pairwise values of θ had to be log transformed (Slatkin, 1993), we added the smallest pairwise θ-value to each estimate so that all logarithms could be calculated. Note that as the different data points are not statistically independent, the number of degrees of freedom is inflated. Therefore, only a nonsignificant correlation coefficient can be interpreted as an indication that there was no relationship. Pairwise values of θ were estimated with FSTAT (Goudet, 1995).
Random effects ANOVA
To estimate how genetic variation is partitioned over the different study sites, regions and time we used a random effects ANOVA with allele frequency as the dependent variable to estimate the variance components for each allele separately. Variance components were divided by the total limiting variance qt(1−qt) (Wright, 1978) of the respective alleles to obtain the proportion of variance. The variance components and total limiting variances were summed over alleles per locus to obtain components and proportions for the five loci. They were then added over the five loci to obtain overall components and proportions. We estimated the variance components and proportions for the following five factors: year, region, region × year, site × year, site × region. Note that we did not estimate the variance component for the main effect site as this factor is nested within region. Statistical significance of the proportion corresponding to each different factor was estimated with a one-sample t-test. With this test we assume that loci have differentiated independently.
REML dendrograms
A restricted maximum likelihood (REML) dendrogram was estimated with the program CONTML in the PHYLIP 3.5 package (Felsenstein, 1993). This method is based on the Brownian motion model and assumes that each locus evolves independently by pure genetic drift. We estimated a REML tree for the complete data set, and for the two years separately. The input order of the study sites was randomized and global rearrangements were applied (Felsenstein, 1993) to obtain the best possible tree topology. Coinciding with the branch lengths, 95 per cent confidence intervals were estimated. A branch length was considered significant if the lower bound was positive. Bootstrapping was not applied because only five loci were scored (Van Dongen, 1995).
Effects of habitat fragmentation
Genetic differentiation
To compare the degree of genetic differentiation among study sites in the continuous forests and the fragmented habitats, we estimated genetic differentiation for each region (or subregion, see above) by means of θ (Weir, 1996) for each locus and overall. Region T was excluded from this analysis because the geographical distances between the sites were much larger compared to the other (sub) regions (Fig. 1). Exact significance levels of the single-locus and overall θ-values were approximated by permuting (1000 permutations) genotypes among subpopulations with the program FSTAT (Goudet, 1995). Overall θ estimates over the five loci were averaged across areas and the median was then compared between the fragmented and continuous (sub)regions with an exact Mann–Whitney U-test in STATXACT (ver. 2.1).
Within-subpopulation genetic diversity
Within-subpopulation genetic diversity was estimated in two ways: the mean number of alleles per locus (MNA) and the expected heterozygosity (Hexp) under Hardy–Weinberg conditions (Nei, 1978). As both estimates, and especially MNA, may be correlated with sample size, statistical analysis needs to take this potential bias into account.
First, we estimated the correlation of the diversity estimates between samples in 1993 and 1994, to determine if the samples could be pooled across years. Genetic diversity was then compared among areas by means of permutation. The permutation test was programmed in Turbo Pascal (ver. 7.0) and set up as follows. We estimated the variance of Hexp and MNA among the different study sites. The exact distribution of these variances under the null hypothesis was approximated by permuting (5000 times) individual genotypes across the different areas, retaining the original sample sizes. The significance level to test if the observed variances were larger than expected under the null hypothesis of no difference, is equal to the proportion of permuted variance estimates that were larger than the observed variance.
To compare overall genetic diversity in the fragmented and continuous (sub)regions we pooled individuals within the different (sub)regions and compared the Hexp. We applied a permutation test (5000 times) to test if Hexp differed significantly between fragmented and continuous (sub)regions.
Finally we related Hexp and MNA to the area and isolation of the different woodlands by means of multiple regression. We added sample size as a covariate to the model to take possible bias into account. Isolation was expressed as the distance to the nearest woodland larger than 10 ha. For study areas larger than 10 ha, isolation was set equal to zero. Initially we explored several measures of isolation, of which this one showed sufficient variation among the different areas and exhibited a smooth distribution without any outliers (see also Van Dongen et al., 1994).
Results
Genetic population structure
Isolation by distance
After log transformation of the pairwise θ values the relationship with geographical distance showed no indication of isolation by distance for either 1993 and 1994 with coefficients of determination lower than 0.01 in both years (P>0.1).
Random effects ANOVA
Proportions of variance of the allele frequencies among the study sites at the different geographical levels were small relative to the total limiting genetic variance. The single-locus values ranged from −0.62–3.5 per cent (Table 2). The largest proportion of genetic variation (2 per cent on average) among the different study sites appeared to be a year by area interaction and differed significantly from zero (t4=5.88, P=0.004). The factors year, region, region×year and region×area were relatively unimportant and did not differ from zero (all P>0.05) (Table 2). These results indicate that the allele frequencies fluctuate from year to year within the different study sites and that there is no or little: (i) overall variation between the two sampling years (no year main effect); (ii) differentiation among regions (no region main effect); (iii) differentiation among areas (no nested area effect (area×region)); and (iv) fluctuation from year to year within the regions (no region by year interaction).
REML dendrograms
Neither samples from the same study site sampled in 1993 and 1994, nor samples from study sites in the same region were clustered together in the REML tree (Fig. 2). Although many branches appeared to be statistically larger than zero, indicating significant differentiation among samples, almost all of these (21 out of 23) were terminal branches. Virtually none of the internal branches differed from zero indicating that the tree structure is unexplained. Thus, although there appeared to be significant differences among some of the samples, there appeared to be no significant structure at a higher level among them. This analysis was also performed for the two sampling years separately and showed the same results (data not shown).
Effects of habitat fragmentation on the distribution of genetic variation
Genetic differentiation
Table 3 summarizes the single and overall (averaged across loci) θ estimates, significance levels and average values of θ for the different (sub)regions in the fragmented and continuous areas. Genetic differentiation was higher in the fragmented areas. For the single-locus θ estimates 13 (37 per cent) out of 35 were statistically significant in the fragmented regions, compared with one (5 per cent) out of 20 in the continuous regions. The latter proportion is exactly equal to the proportion expected by chance alone. Five (71 per cent) out of seven overall θ-values were significant in the fragmented and none out of four in the continuous regions (Table 3). Median θ was larger in the fragmented as compared to the continuous (sub)regions (exact Mann–Whitney U-test, U=3.5, P=0.05). With this test procedure we implicitly assumed that the different overall θ-values were statistically independent. This may not be the case as for (sub)regions H, B2, S1 and P θ estimates for the two sampling years were used. However, a repeated measures analysis could not be performed because of the low number of (sub)regions. Within both sampling years the average θ was larger for the fragmented as compared to the continuous (sub)regions but this difference was not statistically significant for 1993 (U=1, z=1.15, P=0.24) and marginally significant for 1994 (U=0, z=1.85, P=0.06). Note that we applied an asymptotic Mann–Whitney U-test because an exact test cannot detect significant results with the low sample sizes.
Within-subpopulation genetic diversity
For the analysis of genetic diversity we pooled the data of the study sites within the two (sub)regions P and S1 in the continuous areas, as they appeared to show no genetic differentiation (see above).
Hexp was significantly correlated between 1993 and 1994 (r=0.54, d.f.=8, P<0.05). A comparable, yet statistically insignificant, trend was found for MNA (r=0.45, d.f.=8, 0.05<P<0.1). SB was excluded from these analyses because of the small sample size. The correlations indicate that gene diversity differed among areas. However, this analysis did not take the possible effects of variation in sample size into account. After pooling data of 1993 and 1994, Hexp and MNA were still correlated with sample size (Hexp: rSpearman=0.59, N=22, P=0.004; MNA: rSpearman=0.85, N=22, P<0.0001). Note that we applied Spearman rank correlations here as the relationships appeared to be nonlinear. Hexp was significantly correlated with MNA (r=0.70, d.f.=20, P<0.0001). Because sample size was correlated with both Hexp and MNA, we performed a permutation test to investigate if genetic diversity differed among areas. In this way differences in sample sizes among areas were taken into account (see above).
The average of Hexp across areas equalled 0.45 with a variance of 0.0018. This variance was significantly larger than expected by chance (i.e. sampling variance and variation in sample sizes among areas) (permutation test: P=0.0002). The difference in Hexp between the area with the highest (HF: Hexp=0.554) and lowest (LM: Hexp=0.406) equalled 0.148. The average MNA across areas equalled 3.31 with a variance of 0.31. This variance tended to be larger than expected by chance (permutation test: P=0.07). MNA varied between 4 and 2.4.
Hexp for the fragmented (sub)regions was: H=0.503 (N=308), B1=0.496 (N=45), B2= 0.484 (N=412), B3=0.435 (N=45), S2=0.466 (N=56), T=0.431 (N=79); and for the continuous (sub)regions was: S1=0.506 (N=130), P=0.477 (N=175). Average Hexp equalled 0.492 for the continuous and 0.469 for the fragmented (sub)regions. This difference (=0.023) was small, yet statistically significant (permutation test: P=0.03). This analysis was not repeated for MNA as it equalled 4 for all eight (sub)regions.
Hexp was negatively related to woodland isolation (slope=−0.004, SE=0.002, t18=−2.25, P=0.04, Fig. 3) but not area (t18=−0.3, P=0.8), after correction for sample size (log transformed). MNA was not significantly related to woodland isolation or area (P>0.3). Because the relationship between genetic diversity and sample size was nonlinear after log transformation, we also added quadratic terms.
Discussion
The genetic survey of the winter moth presented here reveals counterintuitive results. In contrast to the absence of genetic differentiation at the regional scale (10–40 km), woodland fragments showed significant genetical differences and variation in genetic diversity at the local scale (0.1–3 km). Furthermore, genetic diversity was negatively related to woodland isolation. How can such contradictory results be explained? The different study areas are very alike except for their size and degree of isolation. Therefore, differences in selection pressures are unlikely to explain the observed patterns, although they cannot be ruled out. For FST to be strongly correlated with gene flow there should be an equilibrium between the effects of genetic drift and gene flow. Recent colonizations (Slatkin, 1993), periodic extinctions and recolonizations (Wade & McCauley, 1988), and fluctuations in demographic parameters (Whitlock, 1992) may result in imprecise correlations between FST and Nem. One or a combination of these factors might explain the observed patterns here. The winter moth appears to have been present in Belgium for a long time. Collections of the Royal Belgian Institute of Natural Sciences contain specimens from 1905 and there is no reason to suppose that the winter moth was not already present a long time before that. Furthermore, an examination of 18th century maps showed that there is a high agreement between the woodland configuration of over 200 years ago and now. Oak woods have been present in Belgium for many hundreds of years and the same is probably true for the winter moth. This indicates that there has been sufficient opportunity for an equilibrium to be reached. However, winter moth densities and probably also gene flow are not constant in time. Winter moth densities show strong population cycles of 10–12 years (Altenkirch, 1991). The degree of larval wind dispersal may also vary in time as a result of varying larval hatching/bud burst synchrony (Varley et al., 1973; Van Dongen, 1997). However, the FST value will result in only one average gene flow estimate. In 1993–94, and several years before, winter moth densities were relatively low but increasing, indicating high hatching/bud burst synchrony (Van Dongen et al., 1994; Van Dongen, 1997). As a consequence, larval dispersal may have been relatively low, possibly leading, in combination with relative low effective population sizes, to genetic differentiation and loss of genetic diversity in the fragments. At the regional level effective population sizes are much larger, that at the local level, so that no differentiation at that scale could have evolved during the relatively short period.
As the caterpillars have higher dispersal abilities (see above), geographical factors affecting larval wind dispersal can be expected to influence the population structure. Small and/or isolated woodland patches can thus be expected to receive the fewest immigrants and have the highest probability of becoming extinct. Woodland isolation appeared to be negatively related to genetic diversity, whereas there was no effect for woodland area.
It has been argued that some degree of genetic isolation may be advantageous for the conservation of genetic variation. Computer simulations have shown that overall more genetic diversity may be maintained when the total population is subdivided into a number of subpopulations that diverge from each other. Within-subpopulation genetic diversity will decrease as a result of genetic drift, but because of the overall genetic differentiation among subpopulations, different alleles will be preserved in the different subpopulations resulting in an overall larger genetic diversity (Varvio et al., 1986). However, for the winter moth this does not seem to hold. Genetic diversity is indeed reduced by up to 30 per cent in a number of areas and the genetic differentiation among areas is much lower. Therefore, genetic differentiation among areas does not seem to be able to compensate for the loss of genetic diversity within subpopulations. Indeed, overall genetic diversity was slightly higher in the continuous regions.
References
Altenkirch, W. (1991). Zyklische Fluktuation beim Kleinen Frostspanner (Operophthera brumata L.). Allg Forst Jagd Ztg. 162: 2–7.
Edland, T. (1971). Wind dispersal of the winter moth larvae Operophtera brumata L. (Lep., Geometridae) and its relevance to control measures. Norsk ent Tidsskr. 18: 103–105.
Ellstrand, N. C. and Elam, D. R. (1993). Population genetic consequences of small population size: implications for plant conservation. Ann Rev Ecol Syst. 24: 217–242.
Felsenstein, J. (1993). PHYLIP (Phylogenetic inference package). Version 3.5c. Department of Genetics, University of Washington, Seattle, WA.
Frankham, R. (1995). Conservation genetics. Ann Rev Genet. 29: 305–327.
Goudet, J. (1995). FSTATversion 1.2: a computer program to calculate F-statistics. J Hered. 86: 485–486.
Graf, B., Borer, F., Höpli, H. U., Hohn, H. and Dorn, S. (1995). The winter moth, Operophtera brumata L. (Lep., Geometridae), on apple and cherry: spatial and temporal aspects of recolonization in autumn. J Appl Ent. 119: 295–301.
Harris, H. and Hopkinson, D. A. (1976). Handbook of Enzyme Electrophoresis in Human Genetics. North Holland, Amsterdam.
Hartl, D. L. and Clarke, A. G. (1989). Principles of Population Genetics, 2nd edn Sinauer, Sunderland, MA.
Michalakis, Y., Sheppard, A. W., Noël, V. and Olivieri, I. (1993). Population structure of a herbivorous insect and its host plant on a microgeographic scale. Evolution. 47: 1611–1616.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 89: 583–590.
Skole, D. and Tucker, C. (1993). Tropical deforestation and habitat fragmentation in the Amazon: satellite data from 1978 to 1988. Science. 260: 1905–1910.
Slatkin, M. (1993). Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 47: 264–279.
Stoakley, J. T. (1995). Outbreaks of winter moth, Operophtera brumata L. (Lep., Geometridae) in young plantations of Sitka Spruce in Scotland. Insecticidal control and population assessment using the sex attractant pheromone. Z Ang Ent. 99: 153–160.
Van Dongen, S. (1995). How should we bootstrap allozyme data? Heredity. 74: 445–447.
Van Dongen, S. (1997). Population Structure of the Winter Moth Operophtera brumata in Relation to Local Adaptation and Habitat Fragmentation Ph.D. Dissertation, University of Antwerp, Belgium.
Van Dongen, S., Backeljau, T., Matthysen, E. and Dhondt, A. A. (1994). Effects of forest fragmentation on the population structure of the winter moth Operophtera brumata L. (Lepidoptera, Geometridae). Acta Oecol. 15: 193–206.
Van Dongen, S., Matthysen, E. and Dhondt, A. A. (1996). Restricted male winter moth (Operophtera brumata L.) dispersal among host trees. Acta Oecol. 17: 319–329.
Varley, G. C., Gradwell, G. R. and Hassell, M. P. (1973). Insect Population Ecology. Blackwell Scientific Publications, Oxford.
Varvio, S., Chakraborty, R. and Nei, M. (1986). Genetic variation in subdivided populations and conservation genetics. Heredity. 57: 189–198.
Wade, M. J. and Mccauley, D. E. (1998). Extinction and recolonization: their effects on the genetic differentiation of local populations. Evolution. 42: 995–1005.
Weir, B. S. (1996). Genetic Data Analysis. 2nd edn. Sinauer, Sunderland, MA.
Whitlock, M. C. (1992). Temporal fluctuations in demographic parameters and the genetic variance among populations. Evolution. 46: 608–615.
Wint, W. (1983). The role of alternative host-plants species in the life of a polyphagous moth, Operophtera brumata (Lepidoptera: Geometridae). J Anim Ecol. 52: 439–450.
Wright, S. (1943). Isolation by distance. Genetics. 28: 114–138.
Wright, S. (1978). Evolution and the Genetics of Populations vol. 4 Variability Within and Among Natural Populations. University of Chicago Press, Chicago.
Acknowledgements
S.V.D. is research assistant and E.M. is research associate with the Fund for Scientific Research (Flanders, Belgium). Ellen Sprengers assisted with the field and laboratory work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
van Dongen, S., Backeljau, T., Matthysen, E. et al. Genetic population structure of the winter moth (Operophtera brumata L.) (Lepidoptera, Geometridae) in a fragmented landscape. Heredity 80, 92–100 (1998). https://doi.org/10.1046/j.1365-2540.1998.00278.x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.1998.00278.x
Keywords
This article is cited by
-
Exploring the legacy of goat grazing: signatures of habitat fragmentation on genetic patterns of endemic weevil populations in Northern Isabela Island, Galápagos (Ecuador)
Conservation Genetics (2016)
-
Associations between forest fragmentation patterns and genetic structure in Pfrimer’s Parakeet (Pyrrhura pfrimeri), an endangered endemic to central Brazil’s dry forests
Conservation Genetics (2013)
-
Combining multiple analytical approaches for the identification of population structure and genetic delineation of two subspecies of the endemic Arabian burnet moth Reissita simonyi (Zygaenidae; Lepidoptera)
Conservation Genetics (2012)
-
Landscape Effects on the Genetic Structure of the Ground Beetle Poecilus Versicolor STURM 1824
Biodiversity and Conservation (2006)
-
Genetic diversity of the endangered Chinese endemic herb Primulina tabacum (Gesneriaceae) revealed by amplified fragment length polymorphism (AFLP)
Genetica (2006)