Abstract
The goal of this short review is to consider the interrelated phenomena of phenotypic variation and genetic constraint with respect to plant diversity. The unique aspects of plants, including sessile habit, modular growth and diverse developmental programs expressed at the phytomer level, merit a specific examination of the genetic basis of their phenotypic variation, and how they experience and escape genetic constraint. Numerous QTL studies with wild and domesticated plants reveal that most phenotypic traits are polygenic but vary in the number and effect of the loci contributing, from a few loci of large effects to many with small effects. Further, somatic mutations, developmental plasticity and epigenetic variation, especially gene methylation, can contribute to increases in phenotypic variation. The flip side of these processes, genetic constraint, can similarly be the result of many factors, including pleiotropy, canalization and genetic redundancy. Genetic constraint is not only a mechanism to prevent change, however, it can also serve to direct evolution along certain paths. Ultimately, genetic constraint often comes full circle and is released through events such as hybridization, genome duplication and epigenetic remodeling. We are just beginning to understand how these processes can operate simultaneously during the evolution of ecologically important traits in plants.
Similar content being viewed by others
Introduction
Morphological diversity is generated by a continually shifting balance between variation and constraint. As modern evolutionary biology takes advantage of an enormous wealth of genetic and genomic information, we are gaining greater insight into the complexities and many players in this balance. Understanding genetic controls and constraints on adaptive phenotypic divergence among populations and species has been a long-standing aim in evolutionary biology. The goal of this short review is to consider the interrelated phenomena of phenotype variation and genetic constraint from the particular viewpoint of plant diversification. It is important to consider why we should examine these issues within the specific context of plant evolution. In many ways, plants are masters of phenotypic variation, both within the lifespan of an individual as well as over the course of evolutionary time. This variability is intimately related to their sessile lifestyle, which requires plants to modify both physiology and morphology in response to environmental change of all forms and timescales. From the developmental standpoint, plants can achieve variability through the repetitive and continuous production of the phytomer, the basic module of plant architecture composed of a lateral determinate organ, an axillary meristem, and an internode. Varying developmental programs expressed at the phytomer level generates the enormous morphological diversity observed in plants. These distinct developmental features of plants warrant a focused consideration of their phenotypic variation, and how they experience and escape genetic constraint.
To begin, it is important that we provide definitions for several key terms. Phenotypic variation can be broadly defined as the distribution or range of morphological, phenological, developmental and biochemical traits that are expressed within and among individual taxa. Phenotypic variation is expressed in many forms, from repeated organs on the same plant to phenotypic differences among species or genera. Surprisingly high levels of phenotypic and genetic variation have been described in model, crop species and natural plant populations (reviewed by Alonso-Blanco et al. (2005); Tonsor et al. (2005); Mitchell-Olds and Schmitt (2006)). The causes of phenotypic variance are complex and diverse and include various genetic effects (e.g., Barton and Keigthley, 2002), epigenetic effects (Grant-Downton and Dickenson, 2006), environmental influences affecting metabolism and development termed ‘developmental or phenotypic plasticity’ (e.g., Pigliucci, 2005; West-Eberhard, 2005) and maternal environment effects (e.g., Rossiter, 1998; Roach and Wulff, 1987). In contrast, constraint can be broadly defined as ‘mechanisms or processes that limit the ability of the phenotype to evolve or bias it along particular paths’ (Schwenk and Wagner, 2003). Clearly, constraints can act at different levels (e.g., genetic, developmental and morphological) and can be due to many different underlying causes. Wagner and Misof (1993) have distinguished between generative or developmental constraints vs morphometric constraints. The key to this distinction is that if the former exists, it generally results in the latter, but the simple existence of morphometric constraints may not correlate with constrained developmental processes. There are many known examples of conservation of phenotype without conservation in the underlying genetic or developmental mechanisms, a phenomenon also termed ‘developmental system drift’ (DSD) (True and Haag, 2001). For the purposes of this review, we will focus on generative constraints that are fundamentally genetic in nature. We highlight results from some recent studies to demonstrate how quantitative, developmental and molecular genetics, and genomics are increasing our understanding of phenotypic variation and genetic constraints at many levels. We also consider what implications these advances have for our understanding of the evolution of ecologically important traits.
Genetic effects on phenotypic variation
Phenotypic differences within and among plant populations and species range from subtle to dramatic. Naturally occurring mutations are the primary source for phenotypic variation in plants (Table 1 in Alonso-Blanco et al., 2005). Major questions in evolutionary biology and genomics focus on how natural selection influences standing genetic and phenotypic variation: did selection generate the observed patterns of genetic variation? Does adaptive evolution of novel phenotypes involve few genes of major effect or many of smaller effect? Is phenotypic variation among populations or species governed largely by trait-specific loci that evolve independently or by many pleiotropic loci that can constrain adaptive evolution (reviewed by Orr (2005))? How is differential developmental regulation manifested in phenotypic variation? To address these and related issues, we need to know the location, number, effect size, action and interaction of genes contributing to functional effects and phenotypic differences.
Quantitative trait loci (QTL) mapping studies use the statistical association of molecular markers and phenotypes in segregating progeny to genetically map the genes responsible for variation-specific traits. Although the number of QTL studies with wild plant species is relatively modest and interpretation from these studies are sensitive to various aspects of experimental design and analysis (Beavis, 1994), new insights are emerging. QTL studies indicate that most phenotypic differences among populations or species are polygenic, but that the number and effect size differ across traits and species. Few genes of large effect are typical of cultivated, crop and model plant species, and may be the result of strong selection associated with human domestication of plants (reviewed by Remington and Purugganan (2003)). In wild plants, the results have been more variable; yet, demonstrate the roles of genetic variation and selection in effecting variation in important phenotypic traits. For example, only one to six QTLs, with at least one of large effect, were found to differentiate floral traits of Mimulus lewisii from Mimulus cardinalis (Bradshaw et al., 1998). Further studies with these two Mimulus species substituted Yellow Upper (YUP) alleles (Bradshaw et al., 1998) from the alternate species into the Mimulus lewisii or M. cardinalis genetic background to produce the near isogenic lines (NILs) with the new YUP allele. This single gene substitution significantly altered the floral color phenotype of both species NILs and increased visitation rate of the other species major pollinator (Bradshaw and Schemske, 2003). In contrast, between 11 and 15 QTLs of small effect differentiate mating system traits in Mimulus gutattus from Mimulus nasutus (Fishman et al., 2002), whereas three to seven QTLs of moderate effect differentiate Leptosiphon bicolor from Leptosiphon jepsonii (Goodwillie et al., 2006) in mating system traits. Further, a QTL analysis of leaf shape among F2 progeny derived from an Antirrhinum majus by Antirrhinum charidemi cross identified at least 15 QTLs of small to moderate effect (Langlade et al., 2005). Interestingly the allometric trait space described by the principle component analysis for these two species holds for others in the genus, suggesting that a common genetic control network constrains leaf shape across Antirrhinum species. In addition, microarray studies with crops suggest the power of gene interactions to generate phenotypic variation. For example, gene expression studies with maize demonstrated that 76% of the genes tested were differentially expressed between inbred strains and F2 individuals (Schadt et al., 2003) with significant phenotypic effects.
In addition, large-scale studies of genomic sequence data are providing new insights into the role of selection in maintaining phenotypic and genetic variation at larger phylogenetic scales. Comparisons of the pattern of genome-wide polymorphism to the expected pattern under the null model has implicated natural selection as a causal agent that generated genetic (and phenotypic) differences among ecotypic variants in Arabidopsis (e.g., Nordborg et al., 2005; Schmid et al., 2005). As more model and wild plant genomes are sequenced, the types of analyses that can be conducted, and thus our understanding of the processes that generate phenotypic variation among populations and species will expand.
Epigenetic effects on phenotypic variation
Variation among individuals in the degree and position of DNA methylation, an epigenetic phenomenon broadly observed in plants, produces novel phenotypic variation that can be stably inherited across generations (reviewed by Kalisz and Purugganan (2004)). As methylation of genes can regulate the level of gene expression, these methylated epialleles can increase the range of expression of continuous phenotypic variation within populations (Kalisz and Purugganan, 2004; Rapp and Wendel, 2005). Although we do not currently know the extent to which epigenetic effects generally contribute to phenotypic variation, epialleles have been found in both laboratory and natural populations. For instance, epialleles have been identified in Arabidopsis thaliana for SUPERMAN (SUP), a transcriptional repressor that defines inner floral whorl boundaries (Jacobsen and Meyerowitz, 1997) as well as FWA, homeodomain gene that plays a role in flowering time (Koornneef et al., 1991). For SUP, mutant screens have identified seven loss-of-function alleles, dubbed clark kent (clk), that are heritably hypermethylated, with no nucleotide changes between these clk and wild-type SUP alleles (Jacobsen and Meyerowitz, 1997). In contrast, the gain-of-function epiallele of FWA exhibits stable demethylation of the FWA promoter resulting in ectopic expression of the gene (Soppe et al., 2000). Further, naturally occurring epiallelic variants that increase the range of phenotypic variation have been identified in Linaria vulgaris for a gene-affecting floral symmetry (Cubas et al., 1999), in Zea mays for loci controlling vegetative and seed pigmentation (Chandler et al., 2000) and in A. thaliana for genes regulating pathogen resistance (Stokes et al., 2002; reviewed Kakutani, 2002). The frequency with which methylated epiallelic variation influences ecologically important phenotypic variation in natural plant populations is unknown and awaits further development of molecular genetic methods. Other epigenetic effects in plants are implicated in the releasing of cryptic genetic variation, described below.
Other genetic sources of phenotypic variation
Developmental plasticity (West-Eberhard, 2005) is environmentally mediated alteration in the expression of the underlying genetic architecture of plant traits that can result in changes in phenotypic variation. One example of this comes from A. thaliana grown in controlled environments and under natural spring or autumn germination conditions in the field (Weinig et al., 2002). In this study, QTLs for the timing of bolting (flowering) were found to be specific to the growth environment, suggesting that environmental conditions altered QTL expression. Further, some QTLs of major effect were only seen under ecological field conditions, revealing environment-mediated expression of novel loci and phenotypes. Consistent with this finding, many loci involved with vernalization response have been found to respond to cold temperature, resulting in their ultimate activation or repression (reviewed by Sung and Amasino (2005)). Natural variation in the VRN2 gene of wheat is responsible for differences between winter and spring-flowering varieties (Yan et al., 2004). The sessile nature of plants and the wide range of temperature, light, soil and moisture conditions that they experience can strongly select for condition-dependant gene expression such that developmental plasticity in plants is likely to be common.
Finally, somatic mutation can also contribute to phenotypic variation and adaptive evolution in plants. Axillary meristems of phytomers can differentiate into vegetative or reproductive structures, and unlike animals, somatic mutations in plants can be incorporated into reproductive organs and contribute to population variation (Antolin and Strobeck, 1985).
Surprisingly, even phenotypic traits that are invariant can mask significant genetic variation that can become available when expressed in a different genetic background or environment (Whitehead and Crawford, 2006). In a QTL study, Lauter and Doebley (2002) crossed teosinte to two maize inbred lines to reveal cryptic genetic variation in teosinte for phenotypic traits that are otherwise invariant.
Types and sources of genetic constraint
As defined above, constraint is a broadly applied concept that relates to limitations on the ability of the phenotype to evolve. In some cases, constraint is invoked when a given trait shows little or no variation over evolutionary time. Alternatively, parallelism, the independent evolution of the same feature via the same mechanism, may be owing to certain developmental and/or genetic constraints (Hodin, 2000). Both of these types of constraint may be the product of canalization. The formal definition of canalization is ‘a property of developmental systems which refers specifically to the intrinsic robustness that developmental processes display in response to internal or external perturbations’ (Flatt, 2005). Thus canalization of a developmental process is expected to result in constraint. Typically, constraint is discussed in the context of long evolutionary periods, whereas canalization is often considered at many different levels – within individuals, populations and species. Both intersecting concepts must be applied to specific traits in order to be meaningful (Wagner and Misof, 1993).
Given our earlier discussion of plants as masters of phenotypic variation, it is reasonable to ask, do plants experience genetic constraint in the same way as animals? The answer, of course, is yes, although there are some novel aspects of plant genetics that have significant effects on constraint. In terms of agents of genetic canalization, the chaperone protein Hsp90 has been demonstrated to buffer the expression of cryptic phenotypic variation in both plants and animals (Queitsch et al., 2002; Rutherford and Lindquist, 1998). It has been argued, however, that such molecular chaperones may be particularly important for buffering of environmental responses in sessile organisms such as plants (Sangster and Queitsch, 2005).
Other factors that can produce genetic constraint include pleiotropy, which results when a single gene or allele controls the development of two or more traits (Flatt, 2005). This creates a situation where selection to maintain one trait may constrain the evolvability of another. A particularly well-studied example of pleiotropy in plants is the flowering time locus FLC. In addition to controlling vernalization response (Michaels and Amasino, 1999; Sheldon et al., 2000), this locus has been shown to play roles in water use efficiency (McKay et al., 2003), circadian leaf movements (Swarrup et al., 1999) and nitrogen content (Loudet et al., 2003). FLC is notable for the fact that nonfunctional alleles are common in populations that display a rapid cycling phenotype (Michaels and Amasino, 1999; Sheldon et al., 2000; Schlappi, 2001). This suggests that when a winter annual life cycle is being selectively maintained, the functional FLC allele can constrain other aspects of morphology and physiology. Consistent with this idea, in a detailed study of early- and late-flowering mutants of Arabidopsis, Pouteau et al. (2004) found that the former exhibited a broad release of morphological pleiotropy under short days. One possible explanation for this finding is that most of the factors recruited to function in floral repression are relatively ubiquitous regulators with a high degree of pleiotropy. These loci may impose differential constraints when selection favors different flowering times in natural populations. In another investigation of pleiotropy, Hall et al. (2006) recently used joint-trait QTL mapping analysis of F2 hybrids from two divergent populations of M. guttatus to detect a large number of pleiotropic QTLs. Whereas their individual effects can be modest or small, because pleiotropic QTLs contribute to multiple traits, their overall effect on plant phenotypes can be large, and these pleiotropic QTLs can be a major constraint to phenotypic divergence between populations or species. Importantly, the M. guttatus floral QTLs mapped to nearly the same location in M. nasutus, suggesting a common genetic basis for floral phenotypic divergence within and among this species. This may be evidence of another type of genetic constraint (see discussion of parallelism below).
Pleiotropy can also interact with factors such as genetic redundancy and modularity to produce highly constrained genetic programs. One example of this is the basic floral bauplan, especially the highly consistent positioning of stamens outside of the carpels, which has only one exception across over a quarter of a million species of flowering plants (Martinez and Ramos, 1989). The stability of this organization has multiple contributing components. Pleiotropy exists for several important loci, most notably the transcription factor LEAFY (LFY), which is responsible for the induction of floral meristem identity as well as the activation all of the major floral organ identity genes (Schultz and Haughn, 1991; Weigel et al., 1992; Parcy et al., 1998). The floral organ identity genes themselves often exhibit varying degrees of functional redundancy, with as much as fourfold redundancy observed among the SEPALLATA paralogs (Pelaz et al., 2000; Ditta et al., 2004). This kind of redundancy, which is the result of multiple rounds of gene duplication (Zahn et al., 2005), may be particularly common in plants (Shiu et al., 2005). Another component to the stability of the floral bauplan is the inherent robustness of the genetic interactions among its players. Modeling of these interactions indicates that this pathway constitutes a cohesive genetic module (Espinoza-Soto et al., 2004). Such modularity has long been thought to facilitate canalization within the module, whereas increasing overall evolvability by reducing pleiotropy between modules (Wagner and Altenberg, 1996). Thus, while many aspects of floral organization are invariant (e.g., relative organ position), others remain highly variable (e.g., number of whorls, merosity, fusion between organs) and, moreover, can vary independently of traits such as habit and leaf morphology. As evidence of this type of dissociation, Juenger et al. (2005) found large positive genetic correlations among either flower or leaf traits, but low, nonsignificant genetic correlations between flower and leaf traits. These results support the idea that floral and vegetative modules may experience pleiotropy within the module but evolve independently of one another.
Genetic constraint can shape evolutionary trajectories
Up to this point, we have been considering genetic constraint primarily from the standpoint of factors that limit variation, but another important aspect of genetic constraint is its contribution to parallelism. The repeated, independent recruitment of a particular genetic module to control a homoplastic trait may reflect the fact that the underlying developmental processes are subject to certain constraints (Hodin, 2000). Whereas many examples of this phenomenon have been described from animals (reviewed by Hodin (2000)), several intriguing instances are also known from plants. One of the best documented relates to the genetic control of leaf morphology, one of the most variable traits in plants. The production of compound leaves has evolved dozens of times independently from simple leaved ancestors in angiosperms alone, with additional instances of compound leaf types in other land plants (Bharathan et al., 2002). What almost all of these compound leaves have in common is the expression of type I KNOX genes, which are more typically associated with apical meristems and are absent from the simple leaves of species such as Arabidopsis (Bharathan et al., 2002). It appears that a deeply conserved role in maintaining meristematic identity has predisposed the KNOX pathway to be independently recruited many times to promote indeterminacy in compound leaf primordia. Interestingly, this work has revealed that the distinction between simple and compound leaves is not black and white. KNOX gene expression is also associated with the production of deeply lobed leaves and even simple leaves with toothed margins (Bharathan et al., 2002). A particularly elegant recent study by Hay and Tsiantis (2006) has provided decisive evidence that the KNOX pathway has been independently recruited to promote indeterminacy in leaves and, further, suggests a high degree of lability in the regulatory pathways that repress KNOX genes in a typical simple leaf (Figure 1). This example, as well as others (e.g., Feng et al., 2006) seem to fit into the ‘why reinvent the wheel’ class of parallelism – evolution takes advantage of preexisting pathways that represent ‘easy’ ways to solve similar problems (Hodin, 2000). Other cases of parallelism include the repeated, independent targeting of a particular point in a genetic pathway for modification, which is well known in animals (e.g., Gompel et al., 2005). Examples from plants include shifts in LFY expression as a mechanism for remodeling of inflorescence structure, although the specific nature of the regulatory changes may differ in each case (Yoon and Baum, 2004), and independent downregulation of specific loci in the anthocyanin biosynthetic pathway resulting in multiple reversions to white flowers in Aquilegia (Whittal et al., 2006). Such instances are due to a number of factors including the position of the locus in the pathway and the degree of both pleiotropy and redundancy.
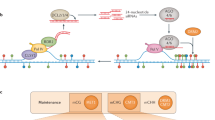
Dramatic diversity in leaf morphology induced in Cardamine hirsute via variations in the expression patterns of endogenous KNOX genes. Silhouettes of rosette (left) and cauline (right) leaves from (a) plants with induced overexpression of KNOTTED1, (b) wild-type plants, and (c) plants with a mutant copy of ASYMMETRIC LEAVES1, a negative regulator of KNOX genes. Arrows denote change in the length of the leaf rachis and arrowheads indicate extra leaflets (Hay and Tsiantis, 2006).
Mechanisms that release genetic constraint
Despite these many sources of constraint, we know that a variety of mechanisms can serve to release even long-standing generative constraints and create phenotypic variation. As discussed above, attenuating the activity of buffering agents such as Hsp90, whether through environmental, epigenetic or genetic effects, can reveal cryptic phenotypic diversity (Sangster and Queitsch, 2005). One notable example of such a response is seen in Linum usitatissimum, common flax, where environmental stress can induce profound genomic changes that increase variability. When these plants are exposed to a variety of inductive environmental conditions, such as different temperature regimes, the genome undergoes heritable, stable rearrangements that result in new morphologies (Cullis, 2005). Another better known mechanism for releasing genetic constraint is genome duplication, which has been shown to have a wide range of immediate and long-term effects (reviewed by Rapp and Wendel (2005); Grant-Downton and Dickinson (2006)). Two important and interacting factors in this regard are epigenetic modification and functional evolution of gene paralogs. Many studies have shown that the former serves to rapidly modify the expression patterns of newly homologous gene copies (e.g., Adams et al., 2003). As this type of selective epigenetic silencing is reversible, it may represent a large amount of latent variation. It is possible that the progeny of such an event could express a high degree of phenotypic variation owing to different patterns of epigenetic modification. Such epigenetic modifications also result in rapid subfunctionalization among homologous gene copies. Although initially epigenetic in nature, this process will serve to maintain multiple gene copies, which can later experience genetic sub- and neofunctionalization. These processes will, in turn, ultimately decrease genetic pleiotropy, further facilitating an increase in evolvability. Interestingly, two important radiations in the angiosperms appear to be associated with genome duplications – the core eudicots and the grasses (Vision et al., 2000; Gaut, 2002; Blanc et al., 2003). A phenomenon that often goes hand in hand with genome duplication is interspecific hybridization, which is particularly common in plants. Plant breeders have used hybridization, likely for millennia, to both increase plant vigor and release variation that is not observed in either parent. Likewise, naturally occurring hybridization has been demonstrated to be important for diversification of plant species (reviewed by Grant-Downton and Dickinson (2006)). The apparently novel variation generated by hybridization is owing to both genetic and epigenetic effects. In terms of the former, transgressive segregation of parental alleles can lead to new combinations not possible in either parental species (Rieseberg et al., 1999). It is clear, however, that epigenetic changes also play a major role as locus activity is remodeled in the progeny genome (Comai et al., 2000). What is intriguing about these mechanisms is that they all have the potential to rapidly change patterns of genetic constraint within the genome, generating large amounts of phenotypic variation for selection to act upon.
Conclusions
Modern biology is rapidly discovering the dynamic nature of phenotypic variation and genetic constraints. Identifying the specific genes represented by QTLs and the function of those genes is a key next step in understanding variation and constraint in the adaptive evolutionary process. This exploration has the potential to reveal the networks of genes in developmental programs responsible for phenotypes, phenotypic integration and constraints. What is particularly fascinating is that many of the processes that generate both genetic variation and constraint are likely to be acting simultaneously in a single genome at any given point. As our understanding of phylogenetic relationships at all levels improves, explicit evolutionary frameworks can be used to examine phenotypic changes from many points of view, including developmental, gene expression (e.g., structural vs regulatory) and ecological (e.g., Whittal et al., 2006). Further, as the genomes of additional model and nonmodel plants are sequenced, comparative genomics will allow additional insights into phenotypic evolution and genetic constraints of ecologically important traits. Among the most promising techniques currently available for elucidating such phenomena are reciprocal transgenics, which permit the dissection of cis- and trans-regulatory changes (e.g., Hay and Tsiantis, 2006), and the application of RNAi-based gene knockdown, which allow endogenous gene functions to be assessed in nonmodel species (Burch-Smith et al., 2004; Hileman et al., 2005). Thus, it will require the application of multiple global approaches and techniques to obtain a full picture of how these sometimes contradictory evolutionary dynamics affect the genome.
References
Adams KL, Cronn R, Percifield R, Wendel JF (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100: 4649–4654.
Alonso-Blanco C, Mendez-Vigo B, Koornneef M (2005). From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. Intl J Dev Biol 49: 717–732.
Antolin MF, Strobeck C (1985). The population genetics of somatic mutations in plants. Am Nat 126: 52–62.
Barton NH, Keigthley PD (2002). Understanding quantitative genetic variation. Nat Rev Gen 3: 11–21.
Beavis WD (1994). The power and deceit of QTL experiments: lessons from comparative QTL studies. Proceedings of the Forty-Ninth Annual Corn Sorghum Industry Research Conference. American Seed Trade Institution: Washington, DC. pp 250–266.
Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860.
Blanc G, Hokamp K, Wolfe KH (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13: 137–144.
Bradshaw HD, Otto KG, Frewen BE, McKay JK, Schemske DW (1998). Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus). Genetics 149: 367–382.
Bradshaw HD, Schemske DW (2003). Allele substitution at a flower color locus produces a pollinator shift in two monkeyflower species (Mimulus). Nature 426: 176–178.
Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP (2004). Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39: 734–746.
Chandler VL, Eggleston WB, Dorweiler JE (2000). Paramutation in maize. Plant Mol Biol 43: 121–145.
Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y et al. (2000). Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12: 1551–1568.
Cubas P, Vincent C, Coen E (1999). An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161.
Cullis CA (2005). Mechanisms and control of rapid genomic changes in flax. Ann Bot 95: 201–206.
Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky M (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940.
Espinoza-Soto C, Padilla-Longoria P, Alvarez-Buylla ER (2004). A gene regulatory network model for cell-fate determination during Arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles. Plant Cell 16: 2923–2939.
Feng XZ, Zhao Z, Tian ZX, Xu SL, Luo YH, Cai ZG et al. (2006). Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci USA 103: 4970–4975.
Flatt T (2005). The evolutionary genetics of canalization. Quar Rev Biol 80: 287–316.
Fishman L, Kelly AJ, Willis JH (2002). Minor quantitative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution 56: 2138–2155.
Gaut BS (2002). Evolutionary dynamics of grass genomes. New Phytol 154: 15–28.
Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB (2005). Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487.
Grant-Downton RT, Dickinson HG (2006). Epigenetics and its implications for plant biology 2. The ‘epigenetic epiphany’: epigenetics, evolution and beyond. Ann Bot 97: 11–27.
Goodwillie C, Ritland C, Ritland K (2006). The genetic basis of floral traits associated with mating system evolution in Leptosiphon (Polemoniaceae): an analysis of quantitative trait loci. Evolution 60: 491–504.
Hall MC, Basten CJ, Willis JH (2006). Pleiotropic quantitative trait loci contribute to population divergence in traits associated with life-history variation in Mimulus guttatus. Genetics 172: 1829–1844.
Hay A, Tsiantis M (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Gen 3: 942–947.
Hileman LC, Drea S, de Martino G, Litt A, Irish VF (2005). Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J 44: 334–341.
Hodin J (2000). Plasticity and constraints in development and evolution. J Exp Zool (Mol Dev Evol) 299: 1–20.
Jacobsen SE, Meyerowitz EM (1997). Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science 277: 1100–1103.
Juenger T, Perez-Perez JM, Bernal S, Micol JL (2005). Quantitative trait loci mapping of floral and leaf morphology traits in Arabidopsis thaliana: evidence for modular genetic architecture. Evol Dev 7: 259–271.
Kalisz S, Purugganan MD (2004). Epialleles via DNA methylation: consequences for plant evolution. Trend Ecol Evol 19: 309–314.
Kakutani T (2002). Epi-alleles in plants: inheritance of epigenetic information over generations. Plant Cell Phys 43: 1106–1111.
Koornneef M, Hanhart CJ, Vanderveen JH (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis-Thaliana. Mol Gen Genet 229: 57–66.
Langlade NB, Feng X, Dransfield T, Copsey L, Hanna AI, Thebaud C et al. (2005). Evolution through genetically controlled allometry space. Proc Natl Acad Sci USA 102: 10221–10226.
Lauter N, Doebley J (2002). Genetic variation for phenotypically invariant traits detected in teosinte: implications for the evolution of novel forms. Genetics 160: 333–342.
Loudet O, Chaillou S, Krapp A, Daniel-Vedele F (2003). Quantitative trait loci analysis of water and anion contents in interaction with nitrogen availability in Arabidopsis thaliana. Genetics 163: 711–722.
Martinez E, Ramos CH (1989). Lacandoniaceae (Triuridales): una nueva familia de Mexico. Ann Mol Bot Gard 76: 128–135.
McKay JK, Richards JH, Mitchell-Olds T (2003). Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Mol Ecol 12: 1137–1151.
Michaels SD, Amasino RM (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956.
Mitchell-Olds T, Schmitt J (2006). Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441: 947–952.
Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H et al. (2005). The pattern of polymorphism in Arabidopsis thaliana. PloS Biol 3: e196.
Orr HA (2005). The genetic theory of adaptation: a brief history. Nat Rev Genet 6: 119–127.
Parcy F, Nilsson O, Busch MA, Lee I, Weigel D (1998). A genetic framework for floral patterning. Nature 395: 561–565.
Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky M (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203.
Pigliucci M (2005). Evolution of phenotypic plasticity: where are we going now? Trends Ecol Evol 20: 481–486.
Pouteau S, Ferret V, Gaudin V, Lefebvre D, Sabar M, Zhao G et al. (2004). Extensive phenotypic variation in early flowering mutants of Arabidopsis. Plant Phys 135: 201–211.
Queitsch C, Sangster TA, Lindquist S (2002). Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624.
Rapp RA, Wendel JF (2005). Epigenetics and plant evolution. New Phytol 168: 81–91.
Remington DL, Purugganan MD (2003). Candidate genes, quantitative trait loci, and functional trait evolution in plants. Int J Plant Sci 164 (Suppl 3): S7–S20.
Rieseberg LH, Archer MA, Wayne RK (1999). Transgressive segregation, adaptation and speciation. Heredity 83: 363–372.
Roach DA, Wulff RA (1987). Maternal effects in plants. Ann Rev Ecol Syst 18: 209–235.
Rossiter MC (1998). The role of environmental variation in parental effects expression. In: Mousseau TA, Fox CW (eds) Maternal Effects As Adaptations. Oxford University Press: New York, NY, USA. pp 112–134.
Rutherford SL, Lindquist S (1998). Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342.
Sangster TA, Queitsch C (2005). The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr Opin Plant Biol 8: 86–92.
Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V et al. (2003). Genetics of gene expression surveyed in maize, moue and man. Nature 422: 297–302.
Schlappi M (2001). RNA levels and activity of FLOWERING LOCUS C are modified in mixed genetic backgrounds of Arabidopsis thaliana. Intl J Plant Sci 162: 527–537.
Schmid KJ, Ramos-Onsins S, Ringys-Beckstein H, Weisshaar B, Mitchell-Olds T (2005). A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169: 1601–1615.
Schultz EA, Haughn GW (1991). LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 3: 771–781.
Schwenk K, Wagner GP (2003). Constraint. In: Hall BK, Olson WM (eds) Keywords and Concepts in Evolutionary Developmental Biology. Harvard University Press: Cambridge, MA. pp 52–60.
Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci USA 97: 3753–3758.
Shiu S-H, Shih M-C, Li W-H (2005). Transcription factor families have much higher expansion rates in plants than in animals. Plant Phys 139: 18–26.
Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M et al. (2000). The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6: 791–802.
Stokes TL, Kunkel BN, Richards EJ (2002). Epigenetic variation in Arabidopsis disease resistance. Genes Dev 16: 171–182.
Sung S, Amasino RM (2005). Remembering winter: toward a molecular understanding of vernalization. Annu Rev Plant Biol 56: 491–508.
Swarrup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino R, Koornneef M et al. (1999). Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J 20: 67–77.
Tonsor SJ, Alonso-Blanco C, Koornneef M (2005). Gene function beyond the single trait: natural variation, gene effects and evolutionary ecology of Arabidopsis thaliana. Plant Cell Environ 28: 2–20.
True JR, Haag ES (2001). Developmental system drift and flexibility in evolutionary trajectories. Evol Dev 3: 109–119.
Vision TJ, Brown DG, Tanksley SD (2000). The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117.
Wagner GP, Altenberg L (1996). Complex adaptations and the evolution of evolvability. Evolution 50: 967–976.
Wagner GP, Misof BY (1993). How can a character be developmentally constrained despite variation in developmental pathways? J Evol Biol 6: 449–455.
Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859.
Weinig C, Ungerer MC, Dorn LA, Kane NC, Toyonaga Y, Halldorsdottir SS et al. (2002). Novel loci control variation in reproductive timing in Arabidopsis thaliana in natural environments. Genetics 162: 1875–1884.
West-Eberhard MJ (2005). Developmental plasticity and the origin of species differences. Proc Nat Acad Sci USA 102: 6543–6549.
Whitehead A, Crawford DL (2006). Variation within and among species in gene expression: raw material for evolution. Mol Ecol 15: 1197–1211.
Whittal JB, Voelckel C, Kliebenstein DJ, Hodges SA (2006). Convergence, constraint and ther role of gene expression during adaptive radiation: floral anthocyanins in Aquilegia. Mol Ecol 15: 4645–4657.
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P et al. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644.
Yoon H-S, Baum DA (2004). Transgenic study of parallelism in plant morphological evolution. Proc Natl Acad Sci USA 101: 6524–6529.
Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL et al. (2005). The evolution of the SEPALLATA subfamily of MADS-Box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169: 2209–2223.
Acknowledgements
This work was supported by NSF Grants DEB 0108208 and 0324764 to SK and IBN 0319103 and EF 0412727 to EK.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kalisz, S., Kramer, E. Variation and constraint in plant evolution and development. Heredity 100, 171–177 (2008). https://doi.org/10.1038/sj.hdy.6800939
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800939
Keywords
This article is cited by
-
Selection criteria for yield potential in a large collection of Vigna radiata (L.) accessions
Euphytica (2020)
-
Comparative sensitivity to gamma radiation at the organismal, cell and DNA level in young plants of Norway spruce, Scots pine and Arabidopsis thaliana
Planta (2019)
-
Quantitative epigenetics and evolution
Heredity (2018)
-
Arabidopsis MSH1 mutation alters the epigenome and produces heritable changes in plant growth
Nature Communications (2015)
-
Genetic and morphological differentiation across a contact zone between two postglacial immigration lineages of Helianthemum nummularium (Cistaceae) in southern Scandinavia
Plant Systematics and Evolution (2015)