Abstract
Evolutionary biologists have long been interested in the processes influencing population differentiation, but separating the effects of neutral and adaptive evolution has been an obstacle for studies of population subdivision. A recently developed method allows tests of whether disruptive (ie, spatially variable) or stabilizing (ie, spatially uniform) selection is influencing phenotypic differentiation among subpopulations. This method, referred to as the FST vs QST comparison, separates the total additive genetic variance into within- and among-population components and evaluates this level of differentiation against a neutral hypothesis. Thus, levels of neutral molecular (FST) and quantitative genetic (QST) divergence are compared to evaluate the effects of selection and genetic drift on phenotypic differentiation. Although the utility of such comparisons appears great, its accuracy has not yet been evaluated in populations with known evolutionary histories. In this study, FST vs QST comparisons were evaluated using laboratory populations of house mice with known evolutionary histories. In this model system, the FST vs QST comparisons between the selection groups should reveal quantitative trait differentiation consistent with disruptive selection, while the FST vs QST comparisons among lines within the selection groups should suggest quantitative trait differentiation in agreement with drift. We find that FST vs QST comparisons generally produce the correct evolutionary inference at each level in the population hierarchy. Additionally, we demonstrate that when strong selection is applied between populations QST increases relative to QST among populations diverging by drift. Finally, we show that the statistical properties of QST, a variance component ratio, need further investigation.
Similar content being viewed by others
Introduction
Most species are subdivided into a network of finite-sized local populations that have become genetically differentiated by the influence of one or more evolutionary processes. Numerous studies have measured levels of genetic differentiation among naturally occurring local populations using different types of molecular and phenotypic data (Turner, 1974; Lessios, 1981; Ritland and Jain, 1984; Schwaegerle et al, 1986; Allegrucci et al, 1987; Baker, 1992; Castric et al, 2001). In each of these studies, genetic divergence among populations was measured; however, determining the relative influence of different evolutionary processes in producing this genetic divergence has been substantially more difficult. This is because different evolutionary processes will often produce similar evolutionary outcomes (Endler, 1986; Slatkin, 1987). For example, spatially variable selection, genetic drift, and/or mutation will all generally result in increased genetic divergence, while uniform selection and/or migration will generally constrain population divergence (Ehrlich and Raven, 1969; Endler, 1986; Lynch, 1986; Slatkin, 1987).
To overcome the difficulty in determining which evolutionary processes are influencing the population differentiation, many studies have integrated both molecular data and quantitative genetic or phenotypic data into studies of population differentiation (see reviews in Reed and Frankham, 2001, Merilä and Crnokrak, 2001, McKay and Latta, 2002). This is done using Wright's (1951) result, which showed the total additive genetic variance in a population under panmixa could be partitioned into within, between, and total components of variation based on Wright's fixation indices (F-statistics). Spitze (1993) formalized this finding and constructed a statistic based on quantitative genetic parameters (QST), which is analogous to Wright's FST. QST measures quantitative genetic divergence among populations and is used in conjunction with FST to assess such data. Estimates of FST from molecular data provide a neutral estimate of genetic differentiation and can be compared with the level of divergence estimated from quantitative genetic data (QST) to determine if the populations are diverging at a rate that is faster, slower, or equal to that expected under neutrality (Lynch and Spitze, 1994). Thus, FST and QST comparisons are generally interpreted in the following manner: if FST is significantly smaller than QST, then spatially variable (ie, disruptive) selection is producing the divergence in quantitative characters among populations; in contrast, if FST is significantly greater than QST, then spatially uniform selection is constraining the divergence in quantitative characters across the landscape and overwhelming the effect of genetic drift. Finally, if FST is not significantly different from QST then the divergence in quantitative characters is potentially occurring at a neutral rate by drift alone. An important caveat about this latter case is that the nonsignificant difference between estimates does not prove that the divergence was the result of genetic drift; rather it demonstrates that the effects of drift and selection are indistinguishable and so natural selection should not be invoked to explain the level of divergence (Merilä and Crnokrak, 2001).
Comparisons of molecular and quantitative genetic variation within and among populations using FST vs QST methodology are increasingly used for testing hypotheses of neutral or adaptive divergence among subpopulations and has been applied in many empirical studies of population differentiation (see references in Merilä and Crnokrak, 2001; McKay and Latta, 2002), as well as being examined theoretically (Latta, 1998; Whitlock, 1999; López-Fanjul et al, 2003). Although this approach has been used in a number of natural systems, this methodology has only been empirically evaluated in a single study involving populations with known evolutionary histories (Porcher et al, 2004) to assess the performance of the FST vs QST comparisons.
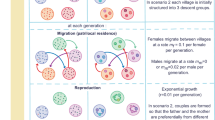
The goal of the present study is to apply FST vs QST comparisons, as they are commonly used in studies of natural populations (Lynch et al, 1999; Morgan et al, 2001; Koskinen et al, 2002; Storz, 2002), to a hierarchically structured laboratory population of house mice (Mus domesticus) with a known evolutionary history: four closed lines selected for high voluntary wheel-running activity and four closed lines of controls random bred with respect to voluntary wheel-running activity. QST values are calculated and compared to previously reported values of FST (Morgan et al, 2003a) in these hierarchical laboratory populations of house mice (Swallow et al, 1998; Figure 1). The hierarchical nature of the population allows the total divergence, FST and QST, to be partitioned into the divergence between selection groups, FGT and QGT, and the divergence among lines within a selection group, FLG and QLG (Smouse and Long, 1987; Weir, 1996). The two different hierarchical levels have different evolutionary processes influencing patterns of genetic differentiation: the effects of selection can be estimated as divergence between selection groups, while the effects of genetic drift can be estimated as the divergence among lines within each selection group. In this study, we estimate QST for wheel-running activity, which has been under strong direct artificial selection (s=0.94 phenotypic standard deviations per generation) for 14 generations (Swallow et al, 1998), and body mass, which has diverged as a result of a negative genetic correlated response to selection (Swallow et al, 1999). Thus, this experimental system models the scenario of a recently derived system of finite populations experiencing strong selection and genetic drift.
If the FST vs QST comparisons perform as expected, then wheel-running activity and body mass should exhibit greater divergence than molecular markers, between the selection groups (ie, FGT<QGT), whereas the level of divergence among the lines within a selection group for wheel-running activity and body mass should be equal to the levels of divergence at molecular markers (ie, FLG=QLG; Figure 1). Additionally, within this single generation, the size of the QGT values among populations diverging as a result of selection should be greater than the QLG values among populations diverging in the absence of selection. Finally, the magnitude of QGT for wheel-running activity should be greater than QGT for body mass because wheel-running activity has been under direct selection whereas body mass has responded in a correlated fashion.
Methods
Evolutionary history
The details of the selection experiment were described previously (Swallow et al, 1998), so only a brief description is provided here. Male and female (112 of each) laboratory house mice (M. domesticus) of the outbred Hsd:ICR strain were purchased from Harlan Sprague–Dawley (Indianapolis, IN, USA). These individuals were paired randomly to produce generation −1. From generation −1, one male and one female were chosen randomly from each litter, and these individuals were paired randomly with the provision of no sib mating. In all, 13 of these pairs were assigned randomly to each of eight closed lines (see Figure 1), and four lines were randomly assigned to each selection group (selection or control). Offsprings from these pairings were designated generation 0, and selection was begun at generation 1. Lines were maintained with 10 pairs per generation through generation 14.
Each generation, mice were weaned from the dams at 21 days of age, weighed, toe-clipped for individual identification and housed, in groups of four, by sex. At 6–8 weeks of age, mice were placed in cages with activity wheels for six consecutive days. The selection trait was average number of wheel revolutions on days 5 plus 6. Within-family selection was used to reduce the effects of inbreeding. In the selected lines, the highest-running male and the highest-running female from each family were chosen to breed, while in control lines, one male and one female from each family were randomly chosen to breed. Breeders were paired randomly within lines for 14 generations, with the condition of no sib mating.
Phenotypic trait collection
Mice from generation 14 were used in this study. At 21 days of age, mice were weaned from dams and placed in individual standard mouse cages. At approximately 7 weeks of age, these mice were then transferred to mouse cages with an attached running wheel (see Swallow et al, 1998). Mice from each line within both selection groups were randomized among the wheels to account for any differences in wheel freeness during the experiment. The mice were allowed to acclimate to the new cages for 4 days, and then the number of revolutions run on days 5 and 6 was recorded for each mouse. The mean number of revolutions run per day on days 5 and 6 was the trait used in the analysis of wheel-running activity. Body mass was also measured, in grams, before and after mice were placed on the wheels; the mean of these body mass measurements was used in the analysis. Following the phenotypic measurements and then pairing to produce generation 15, the mice were killed, and livers were stored at −80°C until used for molecular genetic analysis.
Genotypic data collection
Descriptions of the genotypic data collection can be found in Morgan et al (2003a). Briefly, a subset of the animals from generation 14 was chosen for genotyping by randomly choosing one male and one female from each family. We measured levels of molecular genetic variation using six microsatellite and four allozyme loci. Six highly polymorphic microsatellite loci were chosen from Dietrich et al (1992; D11Mit16, D7Mit18, D13Mit14, D15Mit16) and from Hearne et al (1991; 144, 150). Allozymes genotyped were PGI, PGM, MDH, and 6PGD because they were polymorphic in the base population (Carter et al, 1999). Both the microsatellite and allozyme genotypes were assayed using the same liver tissue. Estimation of the neutral genetic divergence was calculated using all of the polymorphic microsatellite and allozyme markers from generation 14. Although previous studies have suggested that variable mutation rates among different classes of molecular markers may influence the estimation of genetic divergence among populations (Balloux et al, 2000; Hedrick, 1999) we believe the different mutation rates for microsatellite and allozyme loci should have insignificant effects on our estimation of neutral genetic divergence because in these populations, mutations have only been accumulating within lines for 14 generations.
Statistical analysis
Neutral genetic divergence
Morgan et al (2003a) estimated the level of molecular genetic divergence using Wright's F-statistics in a three-level-nested-hierarchical ANOVA with sources of variation between selection groups, between lines within selection group, between individuals within lines, and between alleles within individuals (Weir and Cockerham, 1984; Weir, 1996, Chapter 5). Because of the additional hierarchical level (between selection groups) FST was subdivided into FLG and FGT (Smouse and Long, 1987; Weir, 1996). FLG corresponds to the correlation between randomly chosen gametes within the same line relative to the correlation between randomly chosen gametes within the same selection group. FGT corresponds to the correlation between randomly chosen gametes within the same selection group relative to the correlation between randomly chosen gametes within the total population. For the F-statistics, 95% confidence intervals (CI) were calculated by 10,000 bootstrap replicates of the loci (Manly, 1997). The calculation of F-statistics and 95% CIs were performed using Genetic Data Analysis (Lewis and Zaykin, 2001).
Quantitative genetic divergence
We measured levels of quantitative genetic divergence in two traits, wheel-running activity and body mass. All analyses were performed using sex-corrected measures of wheel-running activity and body mass, because of differences in both traits between the sexes at generation 0 (Swallow et al, 1998). To estimate divergence in quantitative genetic characters we used Wright's (1951) result, which showed the total additive genetic variance for a quantitative character in a population under Hardy–Weinberg equilibrium can be partitioned into within- and between-population components of variation based on his fixation indices (F-statistics) as shown below.

where σ02 is the total additive genetic variance under Hardy–Weinberg equilibrium, and σb2, σw2, and σt2 are the between, within, and total genetic variances respectively. If we assume that local populations are in Hardy–Weinberg equilibrium (FIS=0), as is commonly done in quantitative genetic divergence studies (Spitze, 1993; Yang et al, 1996; Waldmann and Anderson, 1998), and then solve for σ02, the result is a measure for differentiation in quantitative traits that is similar to FST for neutral molecular markers, referred to as QST by Spitze (1993).

The population in this study contains an additional level in the hierarchy (Figure 1), so we partitioned the divergence between lines relative to the total population (QST) into divergence between selection groups relative to the total population (QGT) and divergence among lines relative to each selection group (QLG). To determine the genetic variance within- and among-populations at the different levels in the population's hierarchy the following nested analysis of variance model was used:

where yijkl is the sex-corrected residual phenotypic value of the lth individual within the kth family within jth line within the ith selection group; μ is the overall mean; αi is the ith selection group effect; βj(i) is the jth line effect within the ith selection group; γk(ij) is the kth family effect within the jth line within the ith selection group; and ɛl(ijk) is the within family (residual) variability. Variance components were estimated by equating the observed mean squares from the ANOVA with the expected mean squares (Lynch and Walsh, 1998). Variance components were computed from the sex-corrected residuals using Proc GLM in SAS version 8.1 (SAS Institute, 1994).
The relationship between the observed components of variance from Equation 3 (Vα, Vβ, Vγ) and the causal components of variance (σw2, σb2) needed to calculate quantitative genetic divergence are shown below for divergence between selection groups (QGT) and divergence between lines within selection groups (QLG).

In the calculation of both QGT and QLG the variance among families within lines within selection groups (Vγ) is doubled because individuals in this experiment were full-sibs and thus the among-family component of variance is equal to one-half of the total genetic variance (Falconer and Mackay, 1996). Furthermore, because of the additional hierarchical level in the experiment the genetic variance within populations for the selection groups is equal to the sum of the variance among lines within selection groups (Vβ) plus twice the variance among families within lines within selection groups (Vγ), rather than simply twice the among-family within selection group component of variance, which would ignore the among-line variance.
CIs for Q-statistics were estimated by both nonparametric and parametric bootstrapping (Manly, 1997). For the nonparametric bootstrap individuals were randomly sampled with replacement while maintaining the family sizes and overall structure of the data (ie, individuals per family), and 10 000 nonparametric bootstrap replicates were performed. For the parametric bootstrap, 10 000 data sets were randomly generated from the model (Equation 3) using the estimated parameters computed from the observed data (ie, the standard deviation among the selection groups, lines, and families) and a random number sampled from a normal distribution with mean of zero and standard deviation of 1. From each of the nonparametric and parametric bootstrapped data sets, variance components (Vα, Vβ, Vγ), Q-statistics were estimated and the empirical distributions of the Q-statistics were constructed. The 95% CIs were constructed by the percentile method (Manly, 1997) for both traits measured in this experiment (wheel running, body mass). The calculation of Q-statistics and the 95% CIs was performed in SAS Interactive Matrix Language (IML).
Results
Molecular marker differentiation
Three of the allozyme loci, PGM, MDH, and 6PGD were fixed in all populations and thus were not utilized in the analyses of molecular differentiation. All of the other loci scored appear to be evolving neutrally based on comparisons of observed values of FLG and FGT with expectations under neutrality (Morgan et al, 2003a). This observation of neutrality is of crucial importance if the observed levels of molecular divergence are to be treated as the null hypothesis when testing for adaptive evolution based on divergence in quantitative characters. In addition, the expected proportion of heterozygotes under Hardy–Weinberg equilibrium (ie, FIS=0; see Methods) is satisfied within all lines (Table 1).
As reported by Morgan et al (2003a), the majority of the divergence in molecular markers was attributed to variation among lines within selection groups (FLG), while very little was attributed to variation between the two selection groups (FGT; Table 2). We observed greater levels of divergence among lines within selection groups at all loci, with estimates of FLG ranging from 0.095 to 0.348 and a mean estimate over all loci equal to 0.173, and a 95% CI that did not contain zero (0.115–0.251; Table 2). In contrast, there was very little divergence between selection groups at any of the loci. Estimates of FGT ranged from −0.036 to 0.052 with a mean estimate over all the loci equal to −0.003 (95% CI=–0.028 to 0.014), which was not significantly different from zero (Table 2).
Quantitative trait differentiation
To determine if observed levels of quantitative trait differentiation were significantly different from neutral expectations, both nonparametric and parametric bootstrap approaches were employed (see Methods). For both wheel-running activity and body mass, the nonparametric bootstrap (NPCI) yielded much smaller CIs than were produced from the parametric method (PCI). Both bootstrap approaches were used in this study because a significant bias appeared at some, but not all, hierarchical levels with nonparametric bootstrap. The parametric bootstrap corrected some of these bias issues; however, the parametric approach yields CIs that are substantially larger. Thus we discuss both approaches here and point out that overall interpretation of our results depends on which statistical method is utilized to construct the CI. Our decision to evaluate the significance of the difference between FST and QST statistics based on 95% CIs is generally overly conservative (Manly, 1997); thus significant differences measured between estimates of FST and QST using our methodology should be robust. Alternative methods for evaluating the significance of the difference between estimates of FST and QST could be constructed using Bayesian approaches (Holsinger and Wallace, 2004); however, we have chosen to focus on the bootstrap methodologies as these have been used most commonly to test hypotheses in previous studies.
For wheel-running activity, which was the trait under direct selection (Swallow et al, 1998), the observed levels of quantitative genetic divergence among lines within selection groups (Table 3; QLG=0.1228, 95% NPCI=0.036 to 0.161, 95% PCI=−0.007 to 0.342) was not significantly different from the estimate of divergence based on neutral markers (Table 2; FLG=0.173, 95% CI=0.115–0.251). However, as expected the quantitative genetic divergence between the selection groups was larger than the expectation under neutral divergence. QGT for wheel-running activity was 0.5559 (Table 3; 95% NPCI=0.421 to 0.559, 95% PCI=−0.042 to 0.880), which is greater than our expectation under neutral divergence of FGT=−0.003 (95% CI=−0.028 to 0.014; Table 2). However, our confidence in this FGT vs QGT result for wheel-running activity is dependent on which statistical approach we apply to estimate our CI. With the standard nonparametric bootstrap, we conclude FGT<QGT, which is consistent with our expectation (Figure 1). However with the parametric bootstrap, there is insufficient evidence to reject FGT=QGT.
For body mass, which responded in a negatively correlated fashion to selection for increased wheel-running activity (Swallow et al, 1999; Garland et al, 2002), levels of quantitative genetic divergence among lines within selection groups were significantly less than our expectation of neutral divergence. QLG for body mass was 0.0144 (95% NPCI=0.001 to 0.030, PCI=−0.020 to 0.083; Table 3), which was significantly less than our expected level of divergence under neutrality of FLG=0.173 (95% CI=0.115 to 0.251; Table 2). In contrast, the level of quantitative genetic divergence observed between selection groups was greater than expected under neutrality. QGT for body mass was 0.0641 (95% NPCI=0.0406 to 0.0803, PCI=−0.017 to 0.295), as compared with FGT=−0.003 (−0.028 to 0.014). However, as with wheel-running activity, our ability to assess the significance of this FGT vs QGT result for body mass is dependent on which statistical approach we apply to estimate our CI. With the standard nonparametric bootstrap we conclude FGT<QGT, which is consistent with our expectation (Figure 1). However, with the parametric bootstrap, there is insufficient evidence to reject FGT=QGT.
Discussion
The comparison of estimates of FST and QST is becoming more commonly used in studies of population differentiation (Prout and Baker, 1993; Spitze, 1993; Lynch, 1994; Long and Singh, 1995; Podolsky and Holtsford, 1995; Bonnin et al, 1996; Yang et al, 1996; Waldmann and Anderson, 1998; Lynch et al, 1999; Morgan et al, 2001; Koskinen et al, 2002; Storz, 2002). Although the usefulness of comparisons of FST vs QST in studying potentially adaptive phenotypic divergence appears clear (Merilä and Crnokrak, 2001; McKay and Latta, 2002), such comparisons have not been assessed in a set of populations with known evolutionary histories. Here, we applied FST vs QST comparisons as they are commonly used (Lynch et al, 1999; Morgan et al, 2001; Koskinen et al, 2002; Storz, 2002; Porcher et al, 2004) to a model system composed of eight genetically closed lines of mice from an artificial selection experiment (Swallow et al, 1998). Although this system does not model all the evolutionary processes assumed in most studies of population subdivision in nature (ie, the migration–selection balance), our population does model the scenario of recently derived hierarchical populations experiencing strong selection and significant genetic drift among populations.
At the top level of the population hierarchy (ie, between selection groups), we expected greater levels of divergence in quantitative genetic characters (ie, wheel running and body mass) than in molecular markers because one of the populations has been experiencing strong direct selection for wheel-running activity (s=0.94 phenotypic standard deviations per generation (Swallow et al, 1998)) and correlated responses to selection in body mass (Swallow et al, 1999) for 14 generations. For both wheel-running activity and body mass, we found that the level of divergence in each quantitative character (QGT) was greater than the level of divergence for neutral molecular markers (FGT). However, our confidence in these conclusions was dependent upon which statistical method (nonparametric or parametric) was used to construct our CIs. When the nonparametric method was used, we were able to make the correct evolutionary inference that divergent selection was driving the differentiation between the selection groups. Conversely, when parametric methods were used, equality of FLG and QLG could not be rejected thus leading to the incorrect evolutionary inference that the pattern of divergence between selection groups for quantitative characters is not significantly different from neutral expectations suggesting that the effects of genetic drift and selection are indistinguishable. The discrepancy between parametric and nonparametric estimates of CIs needs further investigation. In particular, if nonparametric methods are producing overly precise statistical confidence in the estimate of QST, then spurious conclusions about the evolutionary phenomenon influencing population differentiation in nature are likely to be inferred. Additionally, Monte Carlo simulations by the second author more generally support these conclusions (Evans, unpublished data), and preliminary analyses suggest that increasing the number of replicate levels (eg, groups, lines, families) results in a reduction of the QST bias.
The level of divergence between selection groups (QGT) was substantially greater for wheel-running activity (Swallow et al, 1998), which was under direct selection, than for body mass, which was diverging by correlated responses to selection (Swallow et al, 1999). This difference is not surprising because direct selection on any trait (assuming that it contains some additive genetic variance) will always produce a more rapid response (divergence) than correlated responses in a second trait unless the two traits are perfectly genetically correlated (Lande and Arnold, 1983).
At the next level in the population hierarchy (ie, lines within selection groups), we expected that the levels of molecular genetic divergence would be equal to the level of quantitative genetic divergence because divergence among lines should generally be the result of genetic drift, given that all lines within a selection group are experiencing similar selection regimes. Although situations do exist where the interaction of selection and drift can increase divergence among populations (Cohan, 1984; Lynch, 1986), this phenomenon does not appear to occur in our study because FLG and QLG are equal for wheel-running activity and QLG is less than FLG for body mass, suggesting that an interaction between selection and genetic drift is not increasing divergence among lines. For wheel-running activity, estimates of FLG and QLG were not significantly different, suggesting the level of divergence in wheel running among lines within selection groups is not consistent with either stabilizing or disruptive selection and implying the divergence at this level is consistent with genetic drift. In contrast, body mass did not match expectations: the level of quantitative genetic divergence among lines within selection groups (QLG) was actually less than the level of molecular genetic divergence (FLG). This result has three possible explanations. First, there might be insufficient additive genetic variance in body mass. However, this is known to be false because Dohm et al (1996, 2001) measured significant narrow-sense heritability for body mass in the base population for the selection study, Swallow et al (1999) showed body mass responded in correlated fashion to selection on wheel running, and we have shown that body mass diverged at the level of the selection group. Second, the finding that QLG is less than FLG may represent a scenario where an FST and QST comparison has failed to yield the correct evolutionary inference in this population with a known evolutionary history. Third, stabilizing selection may in fact be acting to constrain divergence in body mass among lines within selection groups. Although we cannot rule out either the second or third explanations, other results from this system of mice suggest that body mass may be highly constrained in its responses to directional selection and that body mass optima may be essential (Morgan et al, 2003b). Given that we do not have any other evidence suggesting a failure of the methodology at this level of the hierarchy, we favor the stabilizing selection explanation although clearly additional studies are needed to clarify this issue.
In conclusion, our results generally support the ability of FST and QST comparisons to produce the correct evolutionary inference. Our data show that magnitude of the divergence in quantitative characters is greater in populations experiencing strong directional selection (QGT) compared to divergence among populations that are experiencing genetic drift alone (QLT). In addition, the data presented in this study system are similar in nature and design to previously published FST vs QST studies in natural populations (Lynch et al, 1999; Morgan et al, 2001; Koskinen et al, 2002; Storz, 2002). However, although inferences produced from FST vs QST studies generally appear sound, the construction of CIs for QST, a variance component ratio, using standard nonparametric bootstrapping contains a possible significant bias. For wheel-running activity, our estimates of QLG and QGT represent relatively extreme values when compared with the distribution of QLG and QGT produced from our nonparametric bootstrap replicates; that is, the mean of the bootstrap replicates and the point estimates are substantially different at one or more levels in the population hierarchy. This bias is reduced at some (but not all) levels in the population hierarchy when a parametric bootstrap procedure was used; however the parametric procedure resulted in reduced precision of the CIs (ie, the CIs increased). These statistical issues demonstrate that careful consideration of the sampling strategy at each level in a population hierarchy is essential for robust FST vs QST studies.
References
Allegrucci G, Cesaroni D, Sbordoni V (1987). Adaptation and speciation of Dolichopoda cave crickets (Orthoptera, Rhaphidophoridae): geographic variation of morphometric indices and allozyme frequencies. Biol J Linn Soc 31: 151–160.
Baker AJ (1992). Genetic and morphometric divergence in ancestral European and descendent New Zealand populations of chaffinches (Fringilla coelebs). Evolution 46: 1784–1800.
Balloux F, Brunner H, Lugon-Moulin N, Hausser J, Goudet J (2000). Microsatellites can be misleading: an empirical and simulation study. Evolution 54: 1414–1422.
Bonnin I, Prosperi J, Olivieri I (1996). Genetic markers and quantitative genetic variation in Medicago truncatula (Leguminosae): a comparative analysis of population structure. Genetics 143: 1795–1805.
Carter PA, Garland Jr T, Dohm MR, Hayes JP (1999). Genetic variation and correlations between genotype and locomotor physiology in outbred laboratory house mice (Mus domesticus). Comp biochem Phys Part A 123: 155–162.
Castric V, Bonney F, Bernatchez L (2001). Landscape structure and hierarchical genetic diversity in the brook char, Salvelinus fontinalis. Evolution 55: 1016–1028.
Cohan FM (1984). Can uniform selection retard random genetic divergence between isolated conspecific populations? Evolution 38: 495–504.
Dietrich W, Katz H, Lincoln SE, Shin HS, Friedman J, Dracopoli NC et al (1992). A genetic map of the mouse suitable for typing intraspecific crosses. Genetics 131: 423–447.
Dohm MR, Garland Jr T, Hayes JP (1996). Quantitative genetics of sprint running speed and swimming endurance in laboratory house mice (Mus domesticus). Evolution 50: 1688–1701.
Dohm MR, Hayes JP, Garland Jr T (2001). The quantitative genetics of maximal and basal rates of oxygen consumption in mice. Genetics 159: 267–277.
Ehrlich PR, Raven PH (1969). Differentiation of populations. Science 165: 1228–1232.
Endler JA (1986). Natural Selection in the Wild. Princeton University Press, Princeton, NJ.
Falconer DS, Mackay TFC (1996). Introduction to Quantitative Genetics. Longman Press: New York.
Garland Jr T, Morgan MT, Swallow JG, Rhodes JS, Girard I, Belter JG et al (2002). Evolution of a small-muscle polymorphism in lines of house mice selected for high activity levels. Evolution 56: 1267–1275.
Hearne CM, McAleer MA, Love JM, Aitman TJ, Cornall RJ, Ghosh S et al (1991). Additional microsatellite markers for mouse genome mapping. Mamm Genome 1: 273–282.
Hedrick PW (1999). Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution 53: 313–318.
Holsinger KE, Wallace LE (2004). Bayesian approaches for the analysis of population genetic structure: an example from Platanthera leucophaea (Orchidaceae). Mol Ecol 13: 887–894.
Koskinen MT, Haugen TO, Primmer CR (2002). Contemporary fisherian life-history evolution in small salmonid populations. Nature 419: 826–830.
Lande R, Arnold SJ (1983). The measurement of selection on correlated characters. Evolution 37: 1210–1226.
Latta RG (1998). Differentiation of allele frequencies at quantitative trait loci affecting locally adaptive traits. Am Nat 151: 283–292.
Lessios HA (1981). Divergence in allopatry: molecular and morphological differentiation between sea urchins separated by the Isthmus of Panama. Evolution 35: 618–634.
Lewis PO, Zaykin D (2001). Genetic Data Analysis: Computer Program for the Analysis of Allelic Data, Version 1.0 (d16c). Free program distributed by the authors over the internet from http://lewis.eeb.uconn.edu/lewishome/software.html
Long AD, Singh RS (1995). Molecules versus morphology: the detection of selection acting on morphological characters alone a cline in Drosophila melanogaster. Heredity 74: 569–581.
López-Fanjul C, Fernández A, Toro MA (2003). The effect of neutral non-additive gene action on the quantitative index of population divergence. Genetics 164: 1627–1633.
Lynch M (1986). Random drift, uniform selection, and the degree of population differentiation. Evolution 40: 640–643.
Lynch M (1994). Neutral models of phenotypic evolution. In: Real L (ed) Ecological Genetics. Princeton University Press, Princeton, NJ. pp 86–108.
Lynch M, Pfrender M, Spitze K, Lehman N, Allen D, Hicks J et al (1999). The quantitative and molecular genetic architecture of a subdivided species: Daphnia pulex. Evolution 53: 100–110.
Lynch M, Spitze K (1994). Evolutionary genetics of Daphnia. In: Real L (ed) Ecological Genetics. Princeton University Press, Princeton, NJ. pp 109–127.
Lynch M, Walsh B (1998). Genetics and Analysis of Quantitative Traits. Sinauer Associates, Inc.: Sunderland, MA.
Manly BFJ (1997). Randomization, Bootstrap and Monte Carlo Methods in Biology. Chapman & Hall: London.
McKay JK, Latta RG (2002). Adaptive population divergence: markers, QTL and traits. Trends Ecol Evol 17: 285–291.
Merilä J, Crnokrak P (2001). Comparison of genetic differentiation at marker loci and quantitative traits. J Evol Biol 14: 892–903.
Morgan KK, Hicks J, Spitze K, Latta L, Pfrender ME, Weaver CS et al (2001). Patterns of genetic architecture for life-history traits and molecular markers in a subdivided species. Evolution 55: 1753–1761.
Morgan TJ, Garland Jr T, Irwin BL, Swallow JG, Carter PA (2003a). The mode of evolution of molecular markers in populations of house mice under artificial selection for locomotor behavior. J Hered 94: 236–242.
Morgan TJ, Garland Jr T, Carter PA (2003b). Ontogenies in mice selected for high voluntary wheel running activity. I. Mean ontogenies. Evolution 57: 646–657.
Podolsky RH, Holtsford TP (1995). Population structure of morphological traits in Clarkia dudleyana I. Comparison of FST between allozymes and morphological traits. Genetics 140: 733–744.
Porcher E, Giruad T, Goldringer I, Lavinge C (2004). Experimental demonstration of a causal relationship between heterogeneity of selection and genetic differentiation in quantitative traits. Evolution 58: 1434–1445.
Prout T, Baker JSF (1993). F statistics in Drosophila buzzatii: selection, population size, and inbreeding. Genetics 134: 369–375.
Reed DH, Frankham R (2001). How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution 55: 1095–1103.
Ritland K, Jain S (1984). A comparative study of floral and electrophoretic variation with life history variation in Limnanthes alba (Limnanthaceae). Oecologia 63: 243–251.
SAS Institute (1994). SAS/STAT User's Guide Version 6.0. SAS Institute Inc.: Cary, NC.
Schwaegerle KE, Garbutt K, Bazzaz FA (1986). Differentiation among nine population of Phlox. I. Electrophoretic and quantitative variation. Evolution 40: 506–517.
Slatkin M (1987). Gene flow and the geographic structure of natural population. Science 236: 787–792.
Smouse PE, Long JC (1987). A comparative F-statistics analysis of the genetic structure of human populations from lowland South America and Highland New Guinea. In: Weir BS, Goodman MM, Eisen EJ, Namkoong G (eds) Proceedings of the Second International Conference on Quantitative Genetics. Sinauer: Sunderland, MA. pp 32–46.
Storz JF (2002). Contrasting patterns of divergence in quantitative traits and neutral DNA markers: analysis of clinal variation. Mol Ecol 11: 2537–2551.
Spitze K (1993). Population structure in Daphina obtusa: quantitative genetic and allozymic variation. Genetics 135: 367–374.
Swallow JG, Carter PA, Garland Jr T (1998). Artificial selection for increased wheel-running behavior in house mice. Behav Genet 28: 227–237.
Swallow JG, Koteja P, Carter PA, Garland Jr T (1999). Artificial selection for increased wheel-running activity in house mice results in decreased body mass at maturity. J Exp Biol 202: 2513–2520.
Turner BJ (1974). Genetic divergence of death valley pupfish species: biochemical versus morphological evidence. Evolution 28: 281–294.
Waldmann P, Anderson S (1998). Comparison of quantitative genetic variation and allozyme diversity within and between population of Scabiosa canescens and S. columbaria. Heredity 81: 79–86.
Weir B (1996). Genetic Data Analysis II. Sinauer Associates, Inc.: Sunderland, MA.
Weir B, Cockerham C (1984). Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370.
Whitlock MC (1999). Neutral additive genetic variance in a metapopulation. Genet Res 74: 215–221.
Wright S (1951). The genetical structure of populations. Ann Eugenics 15: 323–354.
Yang R, Yeh FC, Yanchuk AD (1996). A comparison of isozyme and quantitative genetic variation in Pinus contorta ssp. latifolia by FST. Genetics 142: 1045–1052.
Acknowledgements
We thank RG Latta, MT Morgan, R Gomulkiewicz, JF Storz, K Holsinger, J Merillä, and two anonymous reviewers for helpful discussions and comments during the preparation of this manuscript. This research was supported by grants from the WSU College of Sciences (to TJM), the WSU School of Biological Sciences Guy Brislawn Fellowship (to TJM), and the National Science Foundation grants DEB-0105079 (to PAC and TJM), and IBN-9728434 and IBN-0212567 (to TG).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Morgan, T., Evans, M., Garland, T. et al. Molecular and quantitative genetic divergence among populations of house mice with known evolutionary histories. Heredity 94, 518–525 (2005). https://doi.org/10.1038/sj.hdy.6800652
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800652
Keywords
This article is cited by
-
QST–FST comparisons: evolutionary and ecological insights from genomic heterogeneity
Nature Reviews Genetics (2013)
-
Divergent evolution of molecular markers during laboratory adaptation in Drosophila subobscura
Genetica (2010)
-
Morphological and microsatellite diversity associated with ecological factors in natural populations of Medicago laciniata Mill. (Fabaceae)
Journal of Genetics (2008)