Abstract
Complementary sex determination in the haplodiploid Hymenoptera leads to the production of inviable or effectively sterile diploid males when diploid progeny are homozygous at the sex-determining locus. The production of diploid males reduces the number of females in a population and biases the effective breeding sex ratio in favor of haploid males. This in turn will reduce the effective population size (Ne) of hymenopteran populations with complementary sex determination relative to the expected reductions due to haplodiploidy alone. The effects of diploid male production on Ne in hymenopterans with complementary sex determination when diploid males are either inviable or effectively sterile are assessed theoretically. In both models, low allelic diversity at the sex locus reduces the Ne of hymenopteran populations, and this effect is largest when diploid males are effectively sterile.
Similar content being viewed by others
Introduction
Wright (1933) demonstrated that the effective population size, the size of a genetically ideal population that has the same rate of heterozygosity loss as an actual population under consideration, for diploids, Ne.d, is

and for haplodiploids, Ne.hd, is

where Nf and Nm are the number of breeding females and males, respectively. The difference between equations (1) and (2) arises due to the lower number of gene copies in haplodiploids, as males are hemizygous at all loci (Hedrick and Parker, 1997). Given equal numbers of breeding males and females, Ne.hd=0.75Ne.d (Crozier, 1976; Hedrick and Parker, 1997).
In the haplodiploid Hymenoptera (ants, bees, wasps, and sawflies), sex is usually determined through the complementary actions of alleles at an autosomal locus (Cook, 1993; Cook and Crozier, 1995; Evans et al, 2004). Complementary sex determination (CSD) is the ancestral sex-determining mechanism in the Hymenoptera (Cook, 1993; Cook and Crozier, 1995), and the CSD locus has recently been characterized in the honeybee Apis mellifera (Beye et al, 2003). Hemizygotes at the sex-determining locus develop from unfertilized eggs into haploid males, while for diploid individuals, heterozygotes and homozygotes develop from fertilized eggs into females and males, respectively (Cook and Crozier, 1995; Beye et al, 2003). Diploid males are either inviable or effectively sterile (Agoze et al, 1994; Cook and Crozier, 1995; Holloway et al, 1999) and, therefore, do not contribute to reproduction in hymenopteran populations. The production of diploid males is essentially equivalent to differential female mortality (Cook and Crozier, 1995). Differential mortality at the end of the parental investment period does not select for changes in the primary sex ratio (Charnov, 1982; Trivers, 1985), and thus the production of diploid males will ultimately bias the effective breeding sex ratio in favor of haploid males (Stouthamer et al, 1992; Cook and Crozier, 1995) in parasitic and mass provisioning species (the majority of bees and solitary wasps mass provision; Bohart and Menke, 1976; Hunt, 1999; Michener, 2000; O'Neil, 2000). This in turn will result in a decrease in Ne of hymenopterans with CSD relative to the expectations of equation (2) for haplodiploids. The effects of diploid male production on Ne in hymenopterans have never been examined.
The effects of diploid male production in hymenopteran populations are assessed theoretically by modifying equation (2) under the following conditions: (1) diploid males are inviable (ie diploid males never emerge as adults) and (2) diploid males are viable but effectively sterile (ie diploid males emerge as adults that are capable of mating, but females mating with diploid males produce triploid progeny). Both scenarios have been documented in the Hymenoptera in both natural and laboratory populations (eg Plowright and Pallett, 1979; Agoze et al, 1994; Duchateau et al, 1994; Cook and Crozier, 1995; Holloway et al, 1999; Krieger et al, 1999; Ayabe et al, 2004; Liebert et al, 2004).
Model
Consider a mass provisioning hymenopteran population producing a proportion of h haploid and 1−h diploid progeny per generation. In haplodiploid populations without CSD, all diploid progeny are expected to be female. However, with CSD, a proportion of diploid progeny will develop into inviable or effectively sterile diploid males that will not contribute to reproduction. Given K effective alleles at the sex-determining locus, the frequency of matched matings, where a mating male and female share a sex allele in common, in a random mating population is (Adams et al, 1977)

Half of the diploid progeny in a matched mating will be homozygous at the sex-determining locus, and thus will develop into diploid males. Assuming that females mate singly, the norm for hymenopterans (eg Eickwort and Ginsberg, 1980; Strassmann, 2001), the proportion of diploids that are male is (Adams et al, 1977; Owen and Packer, 1994)

Given CSD and inviable diploid males, the expected proportion of breeding females is reduced by a factor of 1−Φ. The effective population size for haplodiploids exhibiting CSD with inviable diploid males can then be represented as

where N is the total number of eggs produced per generation. The predictions of this model will also hold if diploid males are viable but never achieve matings, or if they mate but females mating with diploids remate with haploids and utilize the latter's sperm.
When diploid males are effectively sterile (diploid males mate but father only triploid progeny), the proportion of females mating with a diploid male is equivalent to the frequency of males that are diploid, φ:

Only females mating with haploid males will contribute to diploid reproduction, and thus the expected proportion of breeding females is further reduced by a factor of 1−φ when diploid males are effectively sterile and achieve matings. The effective population size for haplodiploids exhibiting CSD with effectively sterile diploid males can then be represented as

Results
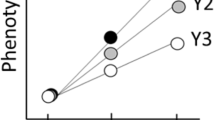
As the effective number of sex alleles approaches infinity, diploid males are seldom produced, and thus equations (6) and (7) converge to equation (2). However, K is finite, and thus the actual proportion of breeding females with CSD, in both models, will always be smaller than the expected proportion of breeding females without CSD, reducing the number of females breeding and biasing the realized sex ratio in favor of haploids. For example, given a primary even sex ratio, where h=(1−h)=0.5 and K=4, the actual proportion of breeding females is reduced by 25% when diploid males are inviable, resulting in an effective breeding sex ratio of 4:3 (haploid males:diploid females). When diploid males are effectively sterile, the same starting parameters result in a 40% reduction in the number of females breeding, and a breeding sex ratio of 5:3.
The effective population size for haplodiploids, and haplodiploids with CSD for different primary sex ratios and numbers of sex alleles are presented in Figure 1. In all cases, the effective population size of hymenopterans with CSD is lower than that expected due to just haplodiploidy. The difference between Ne of populations without CSD (equation (2)) and with CSD (equations (6) and (7)) increases when the effective number of sex alleles is reduced, as expected. The effect of CSD on reducing Ne is stronger when diploid males are effectively sterile versus when they are inviable (Figure 1b versus Figure 1a). When diploid males are inviable (Figure 1a), the magnitude of reduction in Ne increases with male biased primary sex ratios, but the opposite pattern occurs when diploid males are effectively sterile (Figure 1b). The explanation for the former lies in the fact that diploid male production acts to reduce female numbers, the effect of which is most extreme when females are already rare. The latter pattern occurs due to the increased probability of a female mating with a diploid male with female biased primary sex ratios. For example, starting with h=0.1 and K=4, the proportion of females mating with a diploid male is 69% versus only 3% when h=0.9.
Discussion
The effective population size is a parameter of great importance in evolutionary, population, and conservation genetics. For example, Ne controls equilibrium levels of neutral genetic variation, rates of heterozygosity loss, and the relative importance of selection versus drift (Crow and Kimura, 1970; Frankham, 1995; Hedrick, 2000). As many hymenopterans exhibit CSD (Cook and Crozier, 1995), they are expected to have even lower effective population sizes than previously predicted due to haplodiploidy alone. Hymenopteran species usually exhibit lower levels of genetic variation, when compared to diploid insects (reviewed by Packer and Owen, 2001) even when the confounding effects of haplodiploidy and social behavior are removed (Hedrick and Parker, 1997; Packer and Owen, 2001), consistent with the present analysis. Surprisingly low empirical estimates of Ne (≪100) for natural hymenopteran populations (Zayed and Packer, 2001; Antolin et al, 2003; Zayed et al, 2004) also lend support to the view that hymenopterans with CSD have lower Ne than previously expected.
The number of sex alleles in large natural hymenopteran populations usually ranges from 9 to 20 (Cook and Crozier, 1995); however, several studies have documented low allelic diversity at the sex locus (K⩽5) in natural populations (Heimpel et al, 1999; Holloway et al, 1999; Butcher et al, 2000a, 2000b; Carvalho, 2001; Zayed and Packer, 2001; Zayed et al, 2004). Large reductions in Ne relative to equation (2) are thus very likely to occur in small, fragmented, and bottlenecked/introduced populations where allelic diversity at the sex locus is expected to be low (eg Krieger et al, 1999; Zayed and Packer, 2001; Zayed et al, 2004). Similarly, large reductions in Ne are likely to occur in populations of diet specialists expected to have lower allelic diversity at the sex-determining locus than generalist species (Packer et al, 2004). Further, it is theoretically expected that diploid males show higher survival than haploid males, as deleterious recessive alleles are always expressed in the latter. Effective sterility of diploid males is thus likely to be more common than inviability, further increasing the effects of diploid male production on reducing Ne. This is supported by recent reports of triploid females, the product of matings between females and diploid males, in several hymenopteran species (Krieger et al, 1999; Ayabe et al, 2004; reviewed by Liebert et al, 2004). Failure to incorporate diploid male production into population and conservation genetic models may lead to overestimates of Ne, a parameter of great significance to the evolutionary genetics of the Hymenoptera, especially in populations expected to have low allelic diversity at the sex locus.
References
Adams J, Rothman ED, Kerr WE, Paulino ZL (1977). Estimation of the number of sex alleles and queen matings from diploid male frequencies in a population of Apis mellifera. Genetics 86: 583–596.
Agoze ME, Drezen JM, Renalt S, Preiquet G (1994). Analysis of the reproductive potential of diploid males in the wasp Diadromus pulchellus (Hymenoptera: Ichneumonidae). Bull Entomol Res 84: 213–218.
Antolin MF, Ode PJ, Heimpel GE, O'Hara RB, Strand MR (2003). Population structure, mating system, and sex-determining allele diversity of the parasitoid wasp Habrobracon hebetor. Heredity 91: 373–381.
Ayabe T, Hoshiba H, Ono M (2004). Cytological evidence for triploid males and females in the bumblebee, Bombus terrestris. Chromosome Res 12: 215–223.
Beye M, Hasselmann M, Fondrk MK, Page RE, Omholt SW (2003). The gene csd is the primary signal for sexual development in the honey bee and encodes a new SR-type protein. Cell 114: 419–429.
Bohart RM, Menke AS (1976). Sphecid Wasps of the World. University of California Press: Berkeley.
Butcher RDJ, Whitfield WGF, Hubbard SF (2000a). Complementary sex determination in the genus Diadegma (Hymenoptera: Ichneumonidae). J Evol Biol 13: 593–606.
Butcher RDJ, Whitfield WGF, Hubbard SF (2000b). Single-locus complementary sex determination in Diadegma chrysostictos (Gmelin) (Hymenoptera: Ichneumonidae). J Hered 91: 104–111.
Carvalho GA (2001). The number of sex alleles (CSD) in a bee population and its practical importance (Hymenoptera: Apidae). J Hymn Res 10: 10–15.
Charnov EL (1982). The Theory of Sex Allocation. Princeton University Press: Princeton.
Cook JM (1993). Sex determination in the Hymenoptera: a review of models and evidence. Heredity 71: 421–435.
Cook JM, Crozier RH (1995). Sex determination and population biology of the Hymenoptera. Trends Ecol Evol 10: 281–286.
Crow JF, Kimura M (1970). An Introduction to Population Genetics Theory. Harper & Row: New York.
Crozier RH (1976). Counter-intuitive property of effective population size. Nature 262: 384.
Duchateau MJ, Hoshiba H, Velthuis HHW (1994). Diploid males in the bumblebee Bombus terrestris: sex determination, sex alleles and viability. Entomol Exp Appl 71: 263–269.
Eickwort GC, Ginsberg HS (1980). Foraging and mating behavior in Apoidea. Annu Rev Entomol 25: 421–446.
Evans JD, Shearman DCA, Oldroyd BP (2004). Molecular basis of sex determination in haplodiploids. Trends Ecol Evol 19: 1–3.
Frankham R (1995). Effective population size/adult population size ratios in wildlife: a review. Genet Res 66: 95–107.
Hedrick PW (2000). Genetics of Populations 2nd edn. Jones and Bartlett Publishers: Sudbury.
Hedrick PW, Parker JD (1997). Evolutionary genetics and genetic variation of haplodiploids and x-lined genes. Annu Rev Ecol Syst 28: 55–83.
Heimpel GE, Antolin MF, Strand MR (1999). Diversity of sex-determining alleles in Bracon hebetor. Heredity 82: 282–291.
Holloway AK, Heimpel GE, Strand MR, Antolin MF (1999). Survival of diploid males in Bracon sp. near hebetor (Hymenoptera: Braconidae). Ann Entomol Soc Am 92: 110–116.
Hunt JH (1999). Trait mapping and salience in the evolution of eusocial vespid wasps. Evolution 53: 225–237.
Krieger MJB, Ross KG, Chang CW, Keller L (1999). Frequency and origin of triploidy in the fire ant Solenopsis invicta. Heredity 82: 142–150.
Liebert AE, Johnson RN, Switz GT, Starks PT (2004). Triploid females and diploid males: underreported phenomena in Polistes wasps? Insect Soc (in press).
Michener CD. (2000). The Bees of the World. The Johns Hopkins University Press: Baltimore.
O'Neil KM (2000). Solitary Wasps: Behavior and Natural History. Comstock Publishing Associates: Ithaca.
Owen RE, Packer L (1994). Estimation of the proportion of diploid males in populations of Hymenoptera. Heredity 72: 219–227.
Packer L, Owen R (2001). Population genetic aspects of pollinator decline. Conserv Ecol 5: 4 [online] URL: http://www.consecol.org/vol5/iss1/art4.
Packer L, Zayed A, Grixti JC, Ruz L, Owen RE, Vivallo F et al (2004). Conservation genetics of potentially endangered mutualisms: reduced levels of genetic variation in specialist versus generalist bees. Conserv Biol (in press).
Plowright RC, Pallett MJ (1979). Worker-male conflict and inbreeding in bumble bees (Hymenoptera: Apidae). Can Entomol 111: 289–294.
Stouthamer R, Luck RF, Werren JH (1992). Genetics of sex determination and the improvement of biological control using parasitoids. Environ Entomol 21: 427–435.
Strassmann J (2001). The rarity of multiple mating by females of social Hymenoptera. Insect Soc 48: 1–13.
Trivers R (1985). Social Evolution. The Benjamin/Cummings Publishing Company Inc.: Menlo Park.
Wright S (1933). Inbreeding and homozygosis. Proc Natl Acad Sci USA 19: 411–420.
Zayed A, Packer L (2001). High levels of diploid male production in a primitively eusocial bee (Hymenoptera: Halictidae). Heredity 87: 631–636.
Zayed A, Roubik DW, Packer L (2004). Use of diploid male frequency data as an indicator of pollinator decline. Proc R Soc Lond B (Suppl) 271: S9–S12.
Acknowledgements
This work was funded by National Science and Engineering Research Council of Canada scholarships to the author, and a grant to L Packer. I thank L Packer, SI Wright, JS Shore, JC Grixti, and two anonymous reviewers for providing helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zayed, A. Effective population size in Hymenoptera with complementary sex determination. Heredity 93, 627–630 (2004). https://doi.org/10.1038/sj.hdy.6800588
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800588
Keywords
This article is cited by
-
Asymmetrical gene flow between coastal and inland dunes in a threatened digger wasp
Conservation Genetics (2024)
-
Body size and age of drone honeybees (Apis mellifera) affect the structure and characteristics of mating congregations via dispersal
Apidologie (2023)
-
Improving bee health through genomics
Nature Reviews Genetics (2020)
-
Population assessment and foraging ecology of nest aggregations of the rare solitary bee, Eucera longicornis at Gatwick Airport, and implications for their management
Journal of Insect Conservation (2020)
-
High resistance to Sacbrood virus disease in Apis cerana (Hymenoptera: Apidae) colonies selected for superior brood viability and hygienic behavior
Apidologie (2020)