Abstract
In the Rosaceae family, which includes Prunus, gametophytic self-incompatibility (GSI) is controlled by a single multiallelic locus (S-locus), and the S-locus product expressed in the pistils is a glycoprotein with ribonuclease activity (S-RNase). Two populations of flowering cherry (Prunus lannesiana var. speciosa), located on Hachijo Island in Japan’s Izu Islands, were sampled, and S-allele diversity was surveyed based on the sequence polymorphism of S-RNase. A total of seven S-alleles were cloned and sequenced. The S-RNases of flowering cherry showed high homology to those of Prunus cultivars (P. avium and P. dulcis). In the phylogenetic tree, the S-RNases of flowering cherry and other Prunus cultivars formed a distinct group, but they did not form species-specific subgroups. The nucleotide substitution pattern in S-RNases of flowering cherry showed no excess of nonsynonymous substitutions relative to synonymous substitutions. However, the S-RNases of flowering cherry had a higher Ka/Ks ratio than those of other Prunus cultivars, and a subtle heterogeneity in the nucleotide substitution rates was observed among the Prunus species. The S-genotype of each individual was determined by Southern blotting of restriction enzyme-digested genomic DNA, using cDNA for S-RNase as a probe. A total of 22 S-alleles were identified. All individuals examined were heterozygous, as expected under GSI. The allele frequencies were, contrary to the expectation under GSI, significantly unequal. The two populations studied showed a high degree of overlap, with 18 shared alleles. However, the allele frequencies differed considerably between the two populations.
Similar content being viewed by others
Introduction
Self-incompatibility in flowering plants prevents self-fertilization through the rejection of pollen from the same plant. This trait prevents inbreeding. In many species, molecular and classical genetic studies indicate that specificity determination in pollen and style is controlled by a single locus with multiple alleles, the S-locus. In gametophytic self-incompatibility (GSI) systems, pollen specificity is determined by the pollen’s own haploid genotype. GSI is the most common system, and has been claimed in more than 60 families of flowering plants (Kao and McCubbin, 1996).
Molecular studies of the S-locus in three plant families, Solanaceae, Rosaceae and Scrophulariaceae, have shown that the S-locus product in pistils is a basic glycoprotein with ribonuclease (RNase) activity, the S-RNase (Anderson et al, 1986; Sassa et al, 1996; Xue et al, 1996). Richman et al (1995, 1996a) and Richman (2000) directly determined the putative S-genotypes of plants sampled from Solanaceae by sequence polymorphism at the S-RNase locus. Vieira and Charlesworth (2002) revealed sequence variation at the S-locus in Scrophulariaceae. In the Rosaceae, molecular techniques for identifying S-alleles by allele-specific PCR are now being developed for crossing and breeding of fruit cultivars (Sonneveld et al, 2001; Wiersma et al, 2001).
This paper reports on S-allele diversity in two populations of flowering cherry, Prunus lannesiana var. speciosa (Rosaceae). In many previous works on S-allele diversity in natural populations, short-lived herbaceous plants that are physically small have been studied. The flowering cherry, used in the present study, is a forest tree species, and the S-allele diversity in such species might reflect their longevity and large physical size. The objectives of this study were to determine the sequence variations of S-RNases from flowering cherry, and to estimate the number of alleles and the allele frequencies at the S-locus in two natural populations of this species.
Methods
Study sites
P. lannesiana var. speciosa is one of Japan’s flowering cherries, and grows wild throughout the Izu islands (Makino, 1961). The study sites were located on Hachijo Island in the Izu Islands (Figure 1a, b; 33°07′N, 139°48′E), which is about 250 km from Honshu, the main island of Japan. Two populations of flowering cherry were sampled to estimate the number of alleles and the allele frequencies. The populations are about 5 km apart and are called Plot A and Plot B (Figure 1c). In addition, 30 individuals of flowering cherry were preliminarily sampled for PCR amplification in another population, located between Plot A and Plot B. Plot A occupied an area of 0.3 ha and 0.5 ha in Plot B. There were 86 individuals in Plot A and 79 in Plot B. All individuals in each plot were mapped. Individuals were carefully identified by monitoring root connections, because about half the individuals in each plot formed sprouting stumps with several stems. However, the possibility of repeated sampling from the same clone could not be completely excluded. Individuals showing the same S-genotype were considered to derive from the same clone if the distances among them were less than 5 m. The number of individuals in Plot A was thus reduced to 67. No such individuals potentially derived from the same clone were observed in Plot B. However, one individual possessing three S-alleles, a possible triploid, was found. This individual was excluded, and the number of individuals was reduced to 78. Thus, there were 145 individuals in our sample.
Plant materials
Young leaves were collected for DNA extraction and stored at −80°C. Flower buds were also collected, and the styles (with stigma) were excised, immediately frozen in liquid nitrogen and stored at −80°C.
Isolation of DNA and RNA
Total DNA was isolated from about 2 g of leaf tissue from each individual, using the method described by Murray and Thompson (1980). Total RNA was isolated from pistils using the method described by Chang et al (1993).
PCR amplification of S-RNase genes from genomic DNA
To amplify S-RNases from genomic DNA, primers C1-sense (5′-TATTTTCAATTTGTGCAACA-3′) and C5-antisense (5′-CAAAATACCACTTCATGTAA-3′) were designed from conserved domains (C1 and C5) of the P. avium (Sonneveld et al, 2001; Wiersma et al, 2001) and P. dulcis (Ushijima et al, 1998) S-RNases. PCR was performed with 35 cycles at 95°C for 30 s, 50°C for 30 s, 72°C for 1 min with initial denaturing at 95°C for 5 min, and a final extension at 72°C for 5 min. The reaction mixture consisted of 1 × PCR buffer, 200 μM dNTPs, 0.4 μM of each primer, 0.1 U Taq polymerase (Gene Taq, Nippon Gene) and about 10 ng of genomic DNA in a 20 μl reaction volume. Amplified fragments were separated on 1.2% agarose gel and visualized by ethidium bromide staining.
Amplification of cDNA encoding the S-RNase
cDNA was synthesized using RNA PCR Kit (AMV) Ver. 2.1 (TaKaRa Shuzo) with 1 μg of total stylar RNA as a template. First-strand cDNA was synthesized using oligo dT-adaptor primer (including oligo dT and M13–40 sequences; 5′-GTTTTCCCAGTCACGAC-3′). The cDNA for S-RNase was amplified by 3′RACE using C1-sense as the forward primer and reverse primer, which is the cDNA synthesis tag (M13–40 primer; same as above without the dT tail).
Cloning and sequencing
PCR products were cloned using the TOPO TA Cloning kit (Invitrogen). Their sequences were determined by an LI-COR model 4000 DNA sequencer (LI-COR).
Sequence analysis
The nucleotide sequences were analyzed using GENETIX-WIN (Ver. 5.0.2) (SDC) and the deduced amino-acid sequences were aligned using ClustalX 1.8.1 (Thompson et al, 1997). A phylogenetic tree of S-RNases of Rosaceae was constructed by the neighbor-joining method based on multiple alignment. Codon-by-codon alignment of the nucleotide sequences was adjusted manually, and the numbers of synonymous (Ks) and nonsynonymous (Ka) substitutions per site were calculated using DnaSP (Ver. 3.51) (Rozas and Rozas, 1999), with the Jukes–Cantor correction.
Genomic Southern analysis
Total DNAs (1 μg) were digested with three restriction enzymes (BglII, DraI and EcoRV). Genomic Southern analysis was performed as described by Mukai et al (1995) with the following modification. The formamide concentration in the hybridization buffer was 25%, and hybridization was performed at 37°C. The 32P-labeled cDNA fragment of S-RNase was used as a probe. After hybridization, the filters were washed twice at 42°C in 1 × SSC, 0.1% SDS (low stringency), and exposed to Kodak BioMax MS film for at least 5 days at −80°C. The S-genotypes of individuals were determined based on the combination of RFLP patterns detected in three enzyme digestions.
Results
Sequence variation of S-RNases
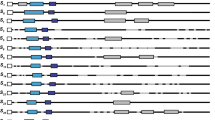
PCR products were detected in 27 of 30 individuals analyzed, of which eight individuals yielded two bands potentially derived from two alleles (Figure 2a). The fragments amplified from genomic DNAs were longer than those expected from the corresponding coding region, and varied in size. This indicates the potential existence of introns, probably varying in size among alleles. The electrophoresis pattern of PCR products showed a fairly close similarity to the RFLP pattern detected in genomic Southern analysis, and the different size fragments corresponded to alleles of types 1–7 determined by RFLP analysis (Figure 2). Therefore, fragments corresponding to different alleles were preferentially selected for cloning. In all, seven sequences were found, and were called types 1–7, respectively. The sequence of type 7 was identical to that of S-RNase from stylar RNA used as the probe for genomic Southern analysis, and matching of these two sequences revealed the presence of one intron. A putative intron of other sequences (types 1–6) was also deduced by comparison with the cDNA sequence of S-RNase. The putative introns differed considerably in size (Table 1). The nucleotide sequences of the exon regions showed high homology (80–90%) to those of the S-RNases previously reported in P. dulcis (Ushijima et al, 1998) and P. avium (Sonneveld et al, 2001; Wiersma et al, 2001). The alignment of the deduced amino-acid sequences is shown in Figure 3. These regions of sequences included the three conserved domains (C2, C3 and RC4) in addition to the C1 and C5 domains used as the PCR primers and a region that is a hyper-variable region (RHV), previously reported in the S-RNases of Rosaceae (Ushijima et al, 1998). The intron position was always in the RHV region, and the RHV was highly variable in the flowering cherry (Figure 3).
PCR amplification of S-RNases from genomic DNA and Southern blotting of restrict enzyme digested genomic DNA. The S-genotype of each individual was determined based on the combination of RFLP patterns detected in BglII, DraI and EcoRV digestions. The types of alleles are indicated at the side of bands probably corresponding to each allele, respectively. (a) Electrophoresis pattern of PCR products amplified from genomic DNA. The band sizes of molecular marker are indicated on the right-hand side. Arrows indicate the size expected from the corresponding coding region. In all, seven alleles were probably amplified, and were called types 1–7, respectively. (b) RFLP pattern detected by genomic Southern analysis. The 32P-labeled cDNA of S-RNase cloned from stylar RNA was used as a probe. Genomic DNAs were digested with DraI. In all, 12 alleles (types 1–12) were found from this sample.
Alignment of the deduced amino-acid sequences of seven S-RNases from flowering cherry. The conserved amino-acid residues are indicated by asterisks. Gaps are indicated by dashes. The C1 and C5 domains used to design the PCR primers, the remaining three conserved domains, C2, C3 and RC4 and one hyper-variable region (RHV), reported in S-RNases of Rosaceae are indicated by underlines and dotted underline, respectively. Arrow indicates the intron position.
A phylogenetic tree was constructed using the neighbor-joining method based on the amino-acid alignment of S-RNases of Rosaceae (data not shown), using three Nicotiana alata S-RNases as outgroups (Figure 4). The S-RNase alleles of Rosaceae fell into two groups: the Malus domestica and Pylus pyrifolia group and a Prunus group, including the cultivars (P. avium and P. dulcis) and the flowering cherry sequences. The seven sequences of the flowering cherry did not cluster together.
Phylogenetic tree of S-RNases in Rosaceae. A phylogenetic analysis was conducted using the neighbor-joining method on the basis of an alignment of 30 S-RNases (data not shown). Sequence data for S-RNases are as follows: S1, S2, S3, S4, S5 and S6 of sweet cherry (P. avium) (Acc. Nos. AB028153, AB010304, AB010306, AB028154, AJ298314, AB010305); Sb, Sc and Sd of almond (Prunus dulcis) (Acc. Nos. AB011469, AB011470, AB011471); S2, S3, S9, S24, S26 and S27 of apple (M. domestica) (Acc. Nos. U12199, U12200, U19793, AF016920, AF016918, AF016919); S1, S2, S3, S4 and S5 of pear (P. pyrifolia) (Acc. Nos. AB002139, AB014073, AB002140, AB014072, AB002141); S2, S3 and S6 of tobacco (N. alata) (Acc. Nos. U08860, U66427, U08861). Arrows indicate the seven S-RNases from flowering cherry determined in this study.
The numbers of synonymous (Ks) and nonsynonymous (Ka) substitutions per site were calculated from pairwise comparisons within and between the seven sequences of flowering cherry, and the nine sequences from Prunus cultivars, S1, S2, S3, S4, S5 and S6 of P. avium (Sonneveld et al, 2001; Wiersma et al, 2001) and Sb, Sc and Sd of P. dulcis (Ushijima et al, 1998). Table 2 shows the results. The mean Ks value of flowering cherry was significantly lower than those of the Prunus cultivars (t test; P<0.01), although there was no difference in the mean Ka value between flowering cherry and Prunus cultivars (t test; P=0.30). Consequently, the mean Ka/Ks ratio for flowering cherry was significantly higher than that for other Prunus cultivars (t test; P<0.01). No excess of nonsynonymous substitutions relative to synonymous substitutions was found in any of the three comparisons. Out of the 21 comparisons of alleles from flowering cherry, only five pairs (2-4, 2-7, 4-7, 5-7, 6-7) showed Ka/Ks ratios slightly greater than 1.0 (Figure 5).
Ka/Ks values for pairwise comparisons of S-RNases from flowering cherry and Prunus cultivars (P. avium and P. dulcis). Ka and Ks values are the numbers of nonsynonymous and synonymous per site. Dotted line, 1:1 ratio of ordinate and abscissa; ▪, comparisons within flowering cherry; □, comparisons within Prunus cultivars (P. avium and P. dulcis); +, comparisons between flowering cherry and Prunus cultivars.
Number of alleles and allele frequency at S-locus
All individuals were analyzed by genomic Southern analysis using a radiolabeled flowering cherry cDNA. Figure 2b is a part of the results, and shows the RFLP pattern of individuals used in the PCR amplification. cDNA for S-RNase used as a probe revealed two or three bands in all individuals, and the RFLPs detected were highly variable. The S-genotypes of individuals were carefully estimated based on the combination of RFLP patterns in three enzyme digestions. Even if some alleles, such as type 2 and type 12 in Figure 2b, were not distinguished in one enzyme digestion, they were distinguished in another enzyme digestion. All individuals showed a heterozygous pattern in the autoradiograms.
In all, 22 different alleles were found; 21 were in Plot A (67 individuals) and 19 were in Plot B (78 individuals). Table 3 shows the frequencies of the different genotypes and alleles in each plot, and Figure 6 shows a histogram of allele frequencies. The two plots were only about 5 km apart (Figure 1c), and showed a high degree of overlap (18 out of a total of 22) in alleles. However, the allele frequencies differed considerably between the two plots, as shown in Figure 6. Nine alleles (type 2, 5, 7, 8, 9, 11, 12, 15 and 17) showed a difference of higher than 0.05 in allele frequency between the two plots. Five alleles (type 6, 16 and three alleles of type ?) occurred at a frequency lower than 0.01 in both plots. The number of alleles estimated by the maximum likelihood method (Paxman, 1963) was almost equal to that observed, not surprisingly, since many individuals were sampled. The assumption of equal allele frequencies, expected under frequency-dependent selection in GSI was tested with Mantel’s (1974) statistic:

where n is the number of alleles observed, r is the number of individuals examined and Cj is the number of times an allele occurs (see also the appendix in Campbell and Lawrence, 1981). The test rejected the hypothesis of equal frequencies for Plot A (χ202 = 98.806, P<0.001) and Plot B (χ182 = 150.258, P<0.001).
Discussion
Determination of S-genotype
This study applied the PCR method using consensus primers as described in the Prunus cultivars (Tao et al, 1999; Sonneveld et al, 2001; Wiersma et al, 2001). This approach was successful in cloning several putative S-RNases and the fragments amplified from the genomic clones of different alleles were distinguished based on the intron length variation. The PCR-based method was thus useful for determining the S-genotypes of individuals. However, the S-alleles other than the seven alleles successfully cloned could not be amplified, and it is not clear how to develop primers for other alleles. Therefore, S-locus genotyping was conducted based on the RFLPs detected in genomic Southern analysis. This approach may overestimate the number of alleles because restriction sites could be outside the coding region of the S-RNase gene. However, this approach was useful for evaluating the variation of many sampled individuals, such as our samples. In addition, the estimated S-genotypes were heterozygous, consistent with the expectation that self-incompatibility prevented the formation of zygotes that are homozygous at the S-locus. Therefore, the estimated genotypes probably reflect the phenotypes of self-incompatibility.
Sequence diversity in S-RNases
The seven sequences amplified from genomic DNA probably corresponded to the alleles identified by genomic Southern analysis, respectively (Figure 2), and one of their sequences was identical to that of S-RNase from stylar RNA. Moreover, all the sequences from flowering cherry have the features and patterns of typical S-RNase, including a single intron in the RHV region (Figure 3), and they were extremely similar to other Prunus S-RNases. These findings support the view that our sequences represent S-RNase.
In the phylogenetic tree, the S-RNases of flowering cherry and Prunus cultivars (P. dulcis and P. avium) clustered together, but they did not form species-specific subgroups. This suggests that the S-RNase of flowering cherry and Prunus cultivars diverged before the divergence of species, as suggested by Ushijima et al (1998).
The Ka/Ks ratio for alleles from flowering cherry did not indicate the positive selection for amino-acid changes, as reported in Physalis crassifolia (Richman et al, 1996b). A recent study of S-RNases in Rosaceae reported that the Ka/Ks ratio for Prunus cultivars (P. avium and P. dulcis) was lower than those for M. domestica and P. pyrifolia. In Malus and Pylus, the Ks and Ka values were 0.24 and 0.20, respectively, giving a Ka/Ks ratio of 0.83 (Ma and Oliveira, 2002). They reported that heterogeneity in nucleotide substitution rates was observed among subfamilies. This study revealed that the Ka/Ks ratio for flowering cherry was higher than that of other Prunus cultivars, and detected a subtle difference among Prunus species. The relatively high Ka/Ks ratio in flowering cherry was due to the Ks value lower than that of other Prunus cultivars. This provides an insight into the evolutionary relationships of Prunus species. Polymorphism at the S-locus is maintained in natural populations for long time periods by balancing selection due to the frequency-dependent advantage of rare alleles (Wright, 1939). Synonymous substitutions that do not result in amino-acid changes are probably subject to drift, while nonsynonymous substitutions that change amino acids are likely to be favored by the balancing selection. Therefore, the relatively low Ks value observed in S-alleles from flowering cherry may be due to genetic drift acting in the populations that maintain these alleles.
S-diversity within and between populations
A total of 22 alleles were found in our sample, and the studied populations were considered to maintain about 20 alleles, which was the mean of the number of alleles for each plot. This number is slightly smaller than those reported for Solanaceae species, whose molecular mechanism of self-incompatibility is similar to that of flowering cherry. The studied populations located in Hachijo Island were geographically isolated, and this might influence the number of alleles maintained in the populations. In previous studies of Solanaceae, although the total number of alleles maintained among two widely separated populations of Solanum carolinense was not much greater than 14 (Richman et al, 1995), populations of P. crassifolia and Lycium andersonii contained 44 alleles (Richman et al, 1996a) and 36 alleles (Richman, 2000), respectively. The population size required to maintain 20 alleles found in the flowering cherry populations was approximately 1500 individuals, assuming mutation rates (to new specificities) of 10−8 per generation (see Figure 2 in Wright, 1939).
Tests of the hypothesis of the equality of allele frequencies, expected under GSI, revealed that the allele frequencies in flowering cherry are significantly uneven, unlike other GSI species, for example, Oenothera organensis where the data fit expectations of equal frequencies (O’Donnell and Lawrence, 1984). We sampled all of the individuals growing in each plot area. Thus, it is likely that individuals sampled in this study were relatively close and this would enhance the potential for the unequal frequency distribution arising from the sampling of relatives. However, individuals possessing the same S-allele were observed at widely scattered locations in each plot area. Therefore, such neighborhood effects are probably negligible. The allele frequencies were not only unequal within the populations but also differed considerably between the two populations. Campbell and Lawrence (1981) proposed two hypotheses to account for such unequal allele frequencies. First, some of the alleles may be subject to an extra effect of selection caused by either a pleiotropic effect of S-genes or close linkage with another gene that affects fitness. Second, unequal allele frequencies may be due to drift and founder effects. Some alleles occurred at a relatively low frequency in both populations studied, and they might be negatively subject to an extra effect of selection. The heterogeneity in allele frequency observed between the two populations was probably caused by drift and founder effect. Flowering cherry is a long-lived species, and, therefore, allele frequency change may be slow. In addition, the density of flowering individuals in the population is low due to their large size. Pollination among nearby and close individuals may thus be common. Together, such features may maintain the complicated allele frequencies within and between flowering cherry populations caused initially by some ‘founder’ or selection event.
References
Anderson MA, Cornish EC, Mau SL, Williams EG, Hoggart R, Atkinson A et al (1986). Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature 321: 38–44.
Campbell JM, Lawrence MJ (1981). The population genetics of the self-incompatibility polymorphism in Papaver rhoeas. II. The number and frequency of S-alleles in a natural population (R106). Heredity 46: 81–90.
Chang S, Puryear J, Cairney J (1993). A simple and efficient method for isolation RNA from pine tree. Plant Mol Biol Rep 11: 113–116.
Kao TH, McCubbin AG (1996). How flowering plants discriminate between self and non-self pollen to prevent inbreeding. Proc Natl Acad Sci USA 93: 12059–12065.
Ma RC, Oliveira MM (2002). Evolutionary analysis of S-RNase genes from Rosaceae species. Mol Genet Genomics 267: 71–78.
Makino T (1961). Makino’s New Illustrated Flora of Japan. Hokuryukan Publishing: Tokyo (in Japanese).
Mantel N (1974). Approaches to a health research occupancy problem. Biometrics 30: 355–362.
Mukai Y, Suyama Y, Tsumura Y, Kawahara T, Yoshimaru H, Kondo T et al (1995). A linkage map for sugi (Cryptomeria japonica) based on RFLP, RAPD, and isozyme loci. Theor Appl Genet 90: 835–840.
Murray MG, Thompson WF (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325.
O’Donnell S, Lawrence MJ (1984). The population genetics of the self-incompatibility polymorphism in Papaver rhoeas. IV. The estimation of the number of alleles in a population. Heredity 33: 159–171.
Paxman GJ (1963). The maximum likelihood estimation of the number of self-sterility alleles in a population. Genetics 48: 1029–1032.
Richman AD (2000). S-allele diversity in Lycium andersonii: implications for the evolution of S-allele age in the Solanaceae. Ann Bot 85: 241–245.
Richman AD, Kao TH, Schaeffer SW, Uyenoyama MK (1995). S-allele sequence diversity in natural populations of Solanum carolinense (Horsenettle). Heredity 75: 405–415.
Richman AD, Uyenoyama MK, Kohn JR (1996a). S-allele diversity in a natural population of Physalis crassifolia (Solanaceae) (ground cherry) assessed by RT-PCR. Heredity 76: 497–505.
Richman AD, Uyenoyame MK, Kohn JR (1996b). Allelic diversity and gene genealogy at the self-incompatibility locus in the Solanaceae. Science 273: 1212–1216.
Rozas J, Rozas R (1999). DnaSP version 3: an integrated program for molecular genetics and molecular evolution analysis. Bioinformatics 15: 174–175.
Sassa H, Nishio T, Kowyama Y, Hirano H, Koba T, Ikehashi H (1996). Self-incompatibility (S) alleles of the Rosaceae encode members of a distinct class of the T2/S ribonuclease superfamily. Mol Gen Genet 250: 547–557.
Sonneveld T, Robbins TP, Boškviæ R, Tobutt KR (2001). Cloning of six cherry self-incompatibility alleles and development of allele-specific PCR detection. Theor Appl Genet 102: 1046–1055.
Tao R, Yamane H, Sugiura A, Murayama H, Sassa H, Mori H (1999). Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNases in sweet cherry. J Am Soc Hortic Sci 124: 224–233.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997). The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882.
Ushijima K, Sassa H, Tao R, Yamane H, Dandekar AM, Gradziel TM, Hirano H (1998). Cloning and characterization of cDNA encoding S-RNases from almond (Prunus dulcis): primary structural features and sequence diversity of the S-RNases in Rosaceae. Mol Genet Genomics 260: 261–268.
Vieira CP, Charlesworth D (2002). Molecular variation at self-incompatibility locus in natural populations of the genera Antirrhinum and Misopates. Heredity 88: 172–181.
Wiersma PA, Wu Z, Zhou L, Hampson C, Kappel F (2001). Identification of new self-incompatibility alleles in sweet cherry (Prunus avium L.) and clarification of incompatibility groups by PCR and sequencing analysis. Theor Appl Genet 102: 700–708.
Wright S (1939). The distribution of self-sterility alleles in populations. Genetics 24: 538–552.
Xue Y, Carpenter R, Dickinson HG, Coen ES (1996). Origin of allelic diversity in Antirrhinum S locus RNase. Plant Cell 8: 805–814.
Acknowledgements
We thank Dr Y Tsumura of the Genome Analysis Laboratory, of the Forestry and Forest Products Research Institute, Japan, for valuable discussions. S Kato thanks her older sister, mother and father for practical help in the field research and the sample collection.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kato, S., Mukai, Y. Allelic diversity of S-RNase at the self-incompatibility locus in natural flowering cherry populations (Prunus lannesiana var. speciosa). Heredity 92, 249–256 (2004). https://doi.org/10.1038/sj.hdy.6800403
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800403
Keywords
This article is cited by
-
Molecular and Evolutionary Characterization of Pollen S Determinant (SFB Alleles) in Four Diploid and Hexaploid Plum Species (Prunus spp.)
Biochemical Genetics (2021)
-
S-genotype identification, genetic diversity and structure analysis of Italian sweet cherry germplasm
Tree Genetics & Genomes (2017)
-
Impact of negative frequency-dependent selection on mating pattern and genetic structure: a comparative analysis of the S-locus and nuclear SSR loci in Prunus lannesiana var. speciosa
Heredity (2012)
-
Genome and population dynamics under selection and neutrality: an example of S-allele diversity in wild cherry (Prunus avium L.)
Tree Genetics & Genomes (2012)
-
Determination S-Genotypes and Identification of Five Novel S-RNase Alleles in Wild Malus Species
Plant Molecular Biology Reporter (2012)