Abstract
Information concerning the area of origin, genetic diversity and possible acquisition of germplasm through hybridisation is fundamental to understanding the evolution, ecology and possible control measures for an introduced invasive plant species. Among the most damaging of alien plants that are invading and degrading native vegetation in the Mascarene Islands of the Indian Ocean is the Tree Privet, Ligustrum robustum. Exact information about the geographic source of introduced material of this species is lacking, in part because Ligustrum is a taxonomically difficult genus. Native material of L. robustum ssp. walkeri from Sri Lanka, L. robustum ssp. robustum from northeastern India, and the closely related L. perrottetii from southern India was compared with introduced material from La Réunion and Mauritius using chloroplast DNA RFLP markers and random amplified polymorphic DNA (RAPDs). Sri Lankan and introduced material was monomorphic for the same cpDNA haplotype that was absent from south and northeast Indian Ligustrum. Sri Lankan and introduced material was also clearly distinguished from Indian Ligustrum by RAPDs. It was concluded that material introduced and established in the Mascarene Islands is derived from the Sri Lankan subspecies L. robustum ssp. walkeri. No geographic structuring of genetic variation within Sri Lanka was detected for this taxon, so the location(s) within Sri Lanka from which introduced material is derived could not be pinpointed. RAPDs indicate that L. robustum ssp. walkeri in Sri Lanka is more similar to south Indian L. perrottetii than to northeast Indian L. robustum ssp. robustum. Moreover, RAPDs showed that introduced material in La Réunion has undergone little or no loss of genetic diversity since introduction. However, there was no evidence that it is introgressed with germplasm from two other alien Ligustrum species present on La Réunion.
Similar content being viewed by others
Introduction
Isolated oceanic tropical islands are remarkable for the high proportion of endemic species, genera and sometimes families among their floras. These floras are, however, subject to serious threats from invading alien species. Introduced plant species now occur in virtually every inhabited part of the world, and a small minority of such plants can become aggressively invasive and displace native species from their habitats (Williamson, 1996; Hollingsworth et al, 1998; Milne and Abbott, 2000; Mooney and Cleland, 2001). The isolated biotas of oceanic tropical islands are particularly vulnerable to invasion by alien species, because they tend to comprise highly adapted specialist rather than generalist species. Generalist species are especially successful in areas of human disturbance, and may also interrupt the intermediate stages of natural succession, such as occur on lava flows on La Réunion in the Mascarene Islands (Macdonald et al, 1991). However, the most damaging aliens are those that invade undisturbed communities, as these can expand their range over time, potentially causing the replacement and extinction of native species. Examples of alien taxa that have invaded native communities on oceanic islands include Myrica faya and Syzygium jambos in Hawaii (Smith, 1989), Cinchona succirubra in the Galapagos Islands (Macdonald et al, 1988), Miconia calvescens in Tahiti (Meyer and Florence, 1996), Cinnomomum zeylandicum in the Seychelles (Freischmann, 1997), and Rubus alceifolius on four Indian Ocean islands (Ansellem et al, 2000).
Among the three islands which comprise the Mascarenes in the Indian Ocean, Mauritius and Rodrigues retain less than 1% of their native vegetation area, and only on La Réunion does a significant area of natural vegetation persist (75 340 ha, ie 30% of the original cover, Strasberg, 1995). Of La Réunion's ∼675 native species, 33% are endemic to the island and a further 22% are endemic to the Mascarenes (Macdonald et al, 1991). The surviving native vegetation on La Réunion is therefore of extreme conservation importance, but is under severe threat from highly invasive introduced species. At least 460 species of vascular plant are naturalised to some extent on La Réunion, and of these, eight have been identified as posing a severe threat to native ecosystems (Macdonald et al, 1991). Among the most noxious of such invasive alien species is the Tree Privet, Ligustrum robustum (Macdonald et al, 1991; Lavergne et al, 1999).
L. robustum was introduced to Mauritius early in the 20th century, and has now penetrated virtually all remaining native vegetation. Together with other aliens, it causes serious degradation of these communities (Macdonald et al, 1991; Lavergne et al, 1999). The introduction of L. robustum to La Réunion occurred much later, probably during the early 1960s and from Mauritian stock (Macdonald et al, 1991; Lavergne et al, 1999). The spread of L. robustum through La Réunion is therefore currently at a correspondingly earlier stage; however, there is ample evidence that it is rapidly expanding its range (Macdonald et al, 1991; Lavergne et al, 1999). For example, between 1989 and 1995 the boundaries of the invading population at Plateau des Chenes above Cilaos have extended from 1000 m altitude to 1300 m altitude: a rate of 50 m altitude per year (Christophe Lavergne, personal communication). Furthermore, L. robustum has become the most serious invading species in the Cilaos forest of La Réunion despite having only been present in the area since 1969 (Macdonald et al, 1991). The advantages held by the species over native competitors on La Réunion have been empirically evaluated, and include superior germination, survivorship and growth rates, possible allelopathy, and shade tolerance (Lavergne et al, 1999). In addition, it produces fruit in large quantities, which might be widely dispersed by birds (Lavergne et al, 1999). Across La Réunion as a whole, Macdonald et al (1991) considered L. robustum the fourth most noxious of all invasive alien species present on the island.
Alien species such as L. robustum that invade undisturbed native communities pose a threat to native biotas which cannot be countered by merely protecting such communities from disturbance, and in many cases cannot be controlled easily or effectively by any form of physical management. Biological control might offer the best method of achieving control of L. robustum on La Réunion, particularly as the species has no close relatives among the indigenous flora. The identification of an effective control organism will be assisted by knowledge of the exact genetic history of the introduced material; specifically, from what geographical source it was introduced and whether it contains genetic material from more than one population or species.
L. robustum is native to the Indo-Malaya region, and is confined to hilly districts, occurring for example between 450 and 2000 m in Sri Lanka (Green, 1987). The typical subspecies is distributed across the region of Asia to the south of the Himalayas, and occurs most commonly in northeastern India. Material from Sri Lanka has been distinguished as subspecies walkeri (Green, 1985) on the basis of small morphological differences. The distribution and taxonomy of many of the Asian species of Ligustrum are incompletely known, largely due to infraspecific variation and the resultant difficulty in identifying material (Green, 1990). In particular, some Ligustrum specimens collected from southern India share characteristics of L. robustum ssp. walkeri and L. perrottetii, the common south Indian species (Green, 1990). This could either reflect natural variation within L. perrottetii, or indicate that L. perrottetii and L. robustum ssp. walkeri actually grade into one another in this area and are not truly distinct.
Material of L. robustum naturalised in Sri Lanka most closely resembles L. robustum ssp. walkeri in morphology. However, based on the above, Ligustrum from India should not be ruled out as the source of the original introduction. Furthermore, the possibility exists that introductions from more than one area have occurred, as for example in British Rhododendron ponticum (Milne and Abbott, 2000) and Bromus tectorum in N America (Novak et al, 1993). Therefore, in the present study, material of Ligustrum from the Mascarene islands was compared, using molecular markers, to that of three native south Asian entities. These were L. robustum ssp. walkeri from Sri Lanka, L. perrottetii from southern India, and L. robustum robustum. The latter taxon was gathered from northeastern India; however, logistical constraints prevented sampling from countries further east.
Introgression has the potential to alter the ecology of introduced species, and potentially render them more invasive (Ellstrand and Schierenbeck, 2000; Milne and Abbott, 2000). Hybrids involving an invasive alien species such as L. robustum might result in the transfer of adaptive traits to the invader through introgression (Rieseberg and Wendel, 1993; Strefeler et al, 1996; Milne and Abbott, 2000), or produce a hybrid lineage more invasive than either parent (Daehler and Strong, 1997; Vila and D'Antonio, 1998; Ellstrand and Schierenbeck, 2000), or that has the potential to be so (Hollingsworth et al, 1998). Two other Ligustrum species, L. sinense and L. ovalifolium, occur as naturalised aliens in La Réunion. Although neither species is yet invasive there, L. sinense is abundantly naturalised in the southeastern USA (Stromayer et al, 1998) and to a lesser extent in Argentina (Montaldo, 1993), and thus might have the potential to become so in the Mascarene Islands. Hybridisation between one of these species and L. robustum, if it occurred, might lead to the evolution of a lineage which is more invasive, and so more destructive to the native ecosystem, than is pure L. robustum. It is therefore important to the study, management and ultimate control of L. robustum in the Mascarenes that such hybridisation events are detected if they occur.
To determine the main source of introduction of L. robustum to the Mascarene Islands, surveys of chloroplast (cp) DNA RFLP and random amplified polymorphic DNA (RAPDs) variation were conducted on material of L. robustum from Sri Lanka, northeastern India, and the Mascarene Islands, and also L. perrottetii from southern India. Material of L. sinense and L. ovalifolium was also examined. RAPD markers were also employed to compare genetic variation within introduced and native material of L. robustum. Reduction of genetic diversity in an introduced population can occur as a result of one or more genetic bottlenecks associated with founder effects (eg Ansellem et al, 2000). Increased genetic diversity in introduced material relative to native material might indicate introgression, whereas direct evidence of this would be provided by the presence of several markers specific to one of the other naturalised species in accessions of L. robustum. RAPDs have previously been employed to demonstrate that introgressive hybridisation has (Daehler and Strong, 1997; Perron and Bousquet, 1997; Hollingsworth et al, 1998) or has not (Miller and Spooner, 1996; Kuehn et al, 1999) occurred between plant species. Moreover, RAPD markers have contributed to the taxonomic understanding of the widespread tropical weed Lantana camara (Scott et al, 1997) and have in some cases been used to indicate the appropriate taxonomic level at which taxa should be recognised (eg Spooner et al, 1997; Paterson and Snyder, 1999).
Materials and methods
Plant collection
All material was collected from the wild by R Shaw, C Lavergne and others (CAB International Bioscience, UK Centre, Silwood Park, Ascot, Berkshire, UK). Material of L. robustum ssp walkeri comprising 41 accessions was sampled from 24 localities covering all five districts in Sri Lanka from which it has been recorded (ie Badulla, Kandy, Monaragalla, Nuwara Eliya and Ratnapura districts; Green, 1987; Table 1). Material of L. perrottetii was gathered from seven localities in the three hilly regions of southern India that are closest to Sri Lanka; that is, Nilgiris, the Palni Hills, and Shevaroys (12 accessions, Table 1). Material of L. robustum ssp. robustum was collected from eight localities in the hills of the Meghalaya region of northeastern India (10 accessions, Table 1). The latter represents only a small sample from the known range of ssp. robustum, and further collections of this taxon would have been made if examination of the introduced material had indicated a genetic affinity with ssp. robustum.
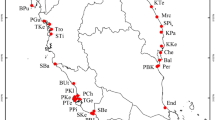
Introduced material from La Réunion was sampled from two upland regions that contain native vegetation remnants that have been invaded by L. robustum, that is, Cirque de Cilaos (24 accessions from seven localities), and Cirque de Salazie (seven accessions, four localities) (Table 1; Figure 1). In addition, specimens were collected from one locality each from the north and south coasts of the island (four accessions, two localities; Table 1; Figure 1). Introduced material was also collected from five localities along an altitude transect on the southwest side of Mauritius (five accessions, five localities).
Map of La Réunion (after Lavergne et al, 1999), showing collection sites of Ligustrum material examined.
Finally, material was collected of two other Ligustrum species, which are naturalised in La Réunion, though to a much more limited extent, that is, material of L. sinense (two localities, four accessions) and L. ovalifolium (one locality, two accessions) (Table 1; Figure 1).
DNA extraction and cpDNA analysis
From each accession, about six young leaves were picked, torn into about four pieces and sealed in bags with ∼25 g of coarse self-indicating silica gel, which was replaced each day until the sample was fully desiccated. Samples were frozen at −20°C as soon as they reached the UK. DNA extraction, DNA purification, and cpDNA RFLP analysis were conducted as described in Milne et al (1999). The pLsC probes from Lactuca sativa, described by Jansen and Palmer (1987), were employed for RFLP analysis.
For cpDNA RFLP analysis, single accessions of Ligustrum, from each of Sri Lanka, northeastern India (Meghalaya) and southern India, were initially examined with the following probes pLsC4 and pLsC6, each in combination with each of 24 restriction enzymes: AluI, BamHI, BclI, BglII, BstEI, ClaI, CfoI, DraI, EcoRI, EcoRV, HaeII, HaeIII, HinfI, HindIII, HpaII, PstI, PvuII, RsaI, SacI, SalI, SmaI, StuI, XbaI, XhoI. When a difference was detected in restriction pattern, then all accessions from India and Sri Lanka (Table 1) were examined to determine if the relevant enzyme/probe combination could distinguish material from one region from that collected from the other two regions. When this was the case, then all samples of material introduced to Mauritius and La Réunion were examined using the same enzyme/probe combination. Within-region cpDNA variation was not sought and might have escaped detection by this method.
RAPD analysis
RAPD reactions were carried out in 25 μl total volumes containing 0.15 μl Taq Polymerase, 1 × Taq reaction buffer, 0.12 mM primer, 0.1 mM DNTP, and 1.5 μl MgCl solution. Primers were selected for use from the following sets: OPA 1-20, OPB 1-20, OPF 1-20, OPG 1-20, OPH 1-20 (Operon Technologies, USA); that is, 100 primers in all. For RAPD reactions, denaturation at 94°C for 3 min was followed by 45 cycles of: 94°C for 30 s, 40°C for 45 s, increased by 0.4°C/s to 72°C, and 90 s at 72°C. A final extension step of 4 min at 72°C was carried out before holding at 5°C. Amplified products were separated in 1.4% agarose gels containing 0.5 μg/μl ethidium bromide, and visualised by UV transillumination.
To test the usefulness of each RAPD primer, three samples of Ligustrum (one from southern India and two from Sri Lanka), were examined and compared in two replicate trials. Primers that produced clear banding patterns for all accessions on both occasions were used in the full survey. These were: OPA2, OPA3, OPA11, OPA13, OPA14, OPA15, OPB5, OPB6, OPB7, OPB8, OPB10, OPB11, OPB12, OPF1, OPF4, OPF9, OPG3, OPG16, OPG17, OPG18, OPH2, OPH3, OPH4, OPH5, OPH7, OPH8, OPH9, OPH13, OPH18.
All clear bands were included in the analyses of variation except for those that occurred in only one extract or in all extracts. Extracts that produced atypical banding patterns for a given locality were examined again to determine if the RAPD bands were genuine or artifacts of contamination. Each band was identified by its position relative to a set of standards (1 kb DNA ladder), and by reference to bands having a constant size within Sri Lankan L. robustum. Whenever there was ambiguity over whether bands in two extracts were of the same size, the two samples were rerun side by side.
A binary data matrix was produced by scoring each RAPD fragment as present or absent from each individual accession. Interindividual relationships in multidimensional space were examined by principal coordinate analysis (PCO) using NTSYSpc2.02i (Rohlf, 1998) on a similarity matrix derived from Jaccard's coefficient, Dj=nxy/n, where nxy is the number of shared markers and n is the total number of markers scored in each pairwise comparison. PCO analysis was conducted on the entire set of material, to determine which native material was most similar to introduced L. robustum material and to detect possible instances of hybridizsation or introgression between the introduced species on La Réunion.
Using the same binary data matrix, a cluster analysis was conducted using the UPGMA method in PAUP* version 4.0b10 (Swofford, 2002), from which a dendrogram was constructed. This tree was bootstrapped with 1000 replicates. It was evident from the results of both the PCO and UPGMA analyses that material from Meghalaya, northeastern India, could be subdivided into two groups, Meghalaya A and B. These were therefore treated as separate entities subsequently.
Further analysis of Ligustrum material from different localities involved comparisons between the following eight ‘populations’: (1) all material from Sri Lanka, (2) all material from southern India, (3) accessions G1–G8 from Meghalaya (‘Meghalaya A’), (4) accessions G9 and G10 from Meghalaya (‘Meghalaya B’), (5) all material of L. robustum from Mauritius, (6) all material of L. robustum from La Réunion, (7) all material of L. sinense from La Réunion, and (8) all material of L. ovalifolium from La Réunion. These populations were included in analyses of molecular variance (AMOVA) of squared Euclidean distances between individuals (Excoffier et al, 1992), using ARLEQUIN 2.001 (Schneider et al, 2000). In addition, AMOVAs were conducted on material from Sri Lanka subdivided into provinces (Table 1) to determine if there was significant RAPD variation between these provinces, and also between these populations and the one from La Réunion.
Shannon's diversity index was used to compare levels of genetic diversity in introduced and native material of L. robustum, by calculating H=Σπ Log 2, where π is the frequency of the ith RAPD band.
Results
Chloroplast DNA variation
Clear differences in restriction fragment pattern were detected between the cpDNA of Sri Lankan L. robustum ssp. walkeri and that of all other native Ligustrum material examined, using the enzymes Cfo1 and Hae2, in combination with pLsC6. A fragment of 1.5 kb in the Cfo1 RFLP profile of south Indian and northeast Indian (Meghalayan) material was replaced by one of 1.3 kb in Sri Lankan material. Similarly, a fragment of 12.2 kb in the Hae2 RFLP profile of Indian material was replaced by two fragments of 10.6 and 1.6 kb in Sri Lankan material. These differences were confirmed in 12 south Indian, eight Meghalayan and 36 Sri Lankan accessions (two Meghalayan and five Sri Lankan accessions did not produce good results). In addition, the accessions of L. sinense and L. ovalifolium both yielded unique species-specific fragment patterns when screened with the Cfo1/pLsC6 combination, but produced profiles identical to Indian L. robustum and L. perrottetii when examined using Hae2/pLsC6. A total of 30 accessions of L. robustum from La Réunion plus all five accessions from Mauritius were examined with both enzyme:probe combinations, and all were found to possess the same cpDNA haplotype as Sri Lankan L. robustum ssp. walkeri.
RAPD variation
The following primers produced amplification products for all (or at least 80% of all) accessions, which included at least one distinct and informative band, and were thus included in the analysis: OPA2, OPA3, OPA13, OPA14, OPB6, OPB7, OPB10, OPB11, OPB12, OPF1, OPF4, OPF9, OPG3, OPG16, OPG17, OPG18, OPH3, OPH4, OPH5, OPH7, OPH8, OPH13, OPH18. A total of 192 polymorphic bands were detected (excluding those present in single accessions only), with up to 13 bands recorded per primer (across all individuals). Although scoring RAPD bands according to distance travelled on an agarose gel permits instances of homoplasy between unconnected bands, using a large set of independent markers should overcome any false impressions produced by homoplasy. Of 65 bands that occurred (at any frequency) in introduced L. robustum, 53 were shared with Sri Lankan material, 39 with south Indian material (L. perrottetii), 24 and 17 with Meghalaya A and B material respectively, 17 with L. sinense and eight with L. ovalifolium. Of the 53 bands present in both Sri Lankan and introduced material, 10 were found in no other material, 12 were otherwise shared with south Indian material only, 12 with south Indian and Meghalayan material only, and 14 with south Indian and/or Meghalayan material and also one or both of the other introduced Ligustrum species, two with Meghalaya only, and three with one of the other introduced species only. Of 12 bands which occurred in introduced L. robustum but not Sri Lankan material, six were found only in introduced L. robustum, three bands (produced by OPA2, OPF1 and OPH8) were shared with L. sinense only, one (produced by OPA13) with L. ovalifolium only, one with both of the other species plus south Indian material, and one with both south Indian and Meghalayan material. Of the bands shared only with either L. sinense or L. ovalifolium, three were detected in single naturalised accessions only, whereas the OPH8 band (shared with L. sinense) was detected in three accessions from Mauritius and two from La Réunion. Two bands (generated by OPF4 and OPB7) that were present at low frequency among Sri Lankan accessions, were absent from all introduced L. robustum, and otherwise were present only in L. sinense. Similarly a band generated by OPA3 was present in 16 native Sri Lankan accessions and was otherwise only present in L. ovalifolium.
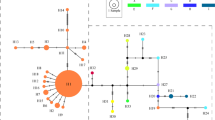
In the PCO analysis of all accessions, the first three PCOs accounted for 29.4, 12.0 and 10.3% of the total variance, respectively. The plot of PCO1 against PCO2 clearly distinguished six discrete groups, of which five were L. sinense, L. ovalifolium, Meghalaya A, Meghalaya B, and south Indian material of Ligustrum (Figure 2). However, material of Ligustrum from Sri Lanka, La Réunion and Mauritius could not be separated and formed a single discrete cluster. There was no indication from this analysis that material of L. robustum introduced to La Réunion was introgressed with germplasm of L. sinense or L. ovalifolium. However, one accession of introduced material, Rc21 (Table 1), possessed a RAPD band that was specific to L. ovalifolium, plus a second that it shared between L. ovalifolium, L. sinense and some south Indian L. perrottetii. Both of these bands are generated by primer OPA13. The presence of these bands in a single accession indicates that introgression cannot entirely be ruled out; however, such bands might also represent symplesiomorphic characters present at low frequency in L. robustum and related species, or indeed nonhomologous bands with similar molecular weights.
The phylogenetic tree produced by UPGMA analysis also supported the subdivision of material examined into the same six groups, each resolved as a distinct clade with >80% bootstrap support (Figure 3). Material from Sri Lanka, La Réunion and Mauritius were grouped together in a single clade with 83% bootstrap support; within this large clade material from the three regions was intermixed, rather than forming discrete subclades. This clade was sister to that comprising southern Indian material (L. perrottetii), with 95% support. There was moderate support (71%) for this clade being sister to one comprising the two entities (A and B) from Meghalaya.
AMOVAs showed that among-population variance was low (11.87%) when material from Sri Lanka, La Réunion and Mauritius populations was compared, but was high (>70%) when introduced material was compared with each of the other native groups (Table 2). AMOVA further showed that there was no significant variation among populations from the three provinces of Sri Lanka, nor between these populations and material from La Réunion (Table 2).
Shannon/Weaver diversity indices (Ho) for material from Sri Lanka, La Réunion and Mauritius were, respectively, 12.084, 10.47 and 6.242, while the diversity index for material from all three localities combined (Ht) was 13.43. Hence diversity is very similar in material from La Réunion and Sri Lanka. The lower diversity index estimated for material from Mauritius might reflect the small sample size (five accessions) and therefore be unreliable.
Discussion
Geographic source of L. robustum introduced to the Mascarene Islands
The cpDNA and RAPD analyses concurred in indicating that introduced material of L. robustum in La Réunion and Mauritius is entirely of Sri Lankan origin. Material of L. robustum ssp. walkeri from Sri Lanka lacks a cpDNA restriction site that is present in Ligustrum material from southern India (L. perrottetii) and Meghalaya, northeastern India (L. robustum ssp. robustum) and also in L. sinense and L. ovalifolium, and therefore has a unique haplotype among the native material examined. The restriction site difference is very likely the apomorphic loss of a Hae2 (cutting sequence (A/G)GCGC(T/C)) cutting site which can also be cut by Cfo1 (cutting sequence GCGC), in all material from Sri Lanka. All examined L. robustum material introduced to La Réunion and Mauritius also lacks this cutting site, and therefore shares the Sri Lankan haplotype.
RAPD analysis revealed clear differences between material of Ligustrum from Sri Lanka, southern India and Meghalaya. However, material from La Réunion, Mauritius and Sri Lanka formed a single cluster according to PCO analysis (Figure 2), and a single clade according to UPGMA analysis (Figure 3). Furthermore, AMOVA tests showed that variation between these populations was low though significant (Table 2). Our data, therefore, indicate unequivocally that the material of L. robustum in the Mascarene Islands is the Sri Lankan taxon, L. robustum ssp. walkeri. To confirm that the source of introduction was Sri Lanka requires a brief re-examination of the possibility that this taxon also occurs on the Indian mainland.
Variation, distribution and relationships among native Asian material of Ligustrum
Ligustrum is a taxonomically difficult genus, in which differences between species may be small, while infraspecific variation is large (Green, 1990). This has led to uncertainty regarding whether L. robustum ssp. walkeri (henceforth, ‘LRW’) is endemic to Sri Lanka, because LRW has previously been recorded also from the hills of southern India. However, the specimens on which those records were based have been tentatively reidentified as L. perrottetii, which is endemic to southern India and is the only Ligustrum species certainly recorded from there (Green, 1990). LRW is found above 450 m in Sri Lanka (Green, 1987), so if it occurs in southern India it would be on uplands only. L. perrottetii is also an upland species, which in the Palni hills area is recorded only above 900 m (Matthew, 1999). In our analysis, material of Ligustrum collected from the three regions of hills in southern India that are closest to Sri Lanka (Palni Hills, Nilgiris, and Shevaroys) were resolved as a single cluster (Figure 2), and a distinct clade (Figure 3), and included no individuals that resembled Sri Lankan material in their marker profiles. Our analysis therefore supports Green's (1990) assertion that LRW does not occur in southern India, and that all material of Ligustrum in this region is referable to a separate taxon (L. perrottetii). Consequently, the source of introduced LRW in the Mascarene Islands can be stated with some confidence to be Sri Lanka.
Among material examined from Meghalaya, northeastern India, there were two distinct genetic entities present (Figures 2 and 3), referred to as Meghalaya A and B. The likely explanation for this is that the two accessions comprising Meghalaya B - that is, G9 and G10 - were actually another Ligustrum species that was misidentified as L. robustum by the collectors. These two accessions differed from G1–8 in having more coriaceous leaves and in growing in more deeply shaded habitats (R Shaw, personal communication). Seven species of Ligustrum besides L. robustum are recorded from Meghalaya, northeastern India, although three are rare and two others easily distinguished from L. robustum by leaf shape (Haridasan and Rao, 1985). Meghalaya B material might in fact be L. lucidum, as this species is frequent in Meghalaya in similar habitats to L. robustum and differs only in minor bract and calyx characters (Haridasan and Rao, 1985).
Among the native material examined, LRW appears to be more similar in RAPD phenotype to L. perrottetii of southern India than it is to L. robustum robustum of Meghalaya, northeastern India, according to the PCO analysis (Figure 2). Moreover, in UPGMA analysis, LRW and L. perrottetii form sister clades that exclude Meghalayan material, with 95% bootstrap support (Figure 3). This apparent similarity conflicts with the taxonomic treatment of Sri Lankan LRW as a subspecies of L. robustum; if LRW is more closely related to L. perrottetii, then it should be elevated to specific status or transferred to a subspecies of L. perrottetii.
Introgression and genetic diversity in introduced Ligustrum
Introduced populations often differ from their progenitor populations in level of genetic diversity. Genetic diversity may be reduced in an introduced population if the population originated from a very small number of founding individuals, and therefore has passed through one or more genetic bottlenecks (eg Ansellem et al, 2000). In the case of LRW in La Réunion, the possibility exists that the material has experienced two such bottlenecks since introduction, as it is thought to have been introduced first to Mauritius, and then from Mauritian stock to La Réunion (Lavergne et al, 1999).
According to Shannon–Weaver diversity indices calculated from our RAPD data, however, there has not been a major loss of diversity in the material in La Réunion (Ho=10.47) relative to native material in Sri Lanka (Ho=12.08). Unfortunately, too few accessions were available from Mauritius to examine meaningfully the diversity of material in this island. In the case of R. alceifolius, which like LRW was introduced first to another island (Madagascar) and from there to La Réunion, there was a dramatic drop in genetic diversity between native and La Réunion populations, such that the latter only contained a single genotype for the markers used (Ansellem et al, 2000). Such a loss of diversity is an expected consequence of founder events associated with plant introductions, but it does not appear to have occurred in LRW on La Réunion. Assuming that the record of LRW being originally introduced to La Réunion from Mauritius is accurate, two hypotheses could explain the relatively high genetic diversity within the La Réunion population. First, both the initial introduction of LRW to Mauritius and the subsequent introduction to La Réunion might have involved large numbers of individuals, probably taken from multiple populations. Second, the original introduction from Mauritius to La Réunion might have been supplemented by new material imported directly from Sri Lanka. The second hypothesis would be supported if further studies revealed lower diversity in populations of LRW in Mauritius than in those in La Réunion.
The genetic diversity of an introduced species can also be increased by introgression (Abbott, 1992; Ellstrand and Schierenbeck, 2000; Milne and Abbott, 2000). In the case of a noxious invading alien, introgression could accelerate the spread of the species or expand the range of habitats it can invade, and might also decrease the effectiveness of control measures employed. The potential for introgression among Ligustrum species certainly exists in the Mascarene islands, where two species besides LRW are also naturalised, though to a lesser extent.
If accessions of introduced LRW had been subject to introgression, these would appear displaced from the main cluster in a PCO analysis of RAPD data. However, this was not found to be the case in the present study (Figure 1). Nevertheless, one accession, Rc21, was found to share one RAPD band uniquely with L. ovalifolium, and another, generated by the same primer, with both L. ovalifolium and L. sinense plus some south Indian L. perrottetii. Given its presence in at least three Ligustrum species, the latter band might be symplesiomorphic among Ligustrum species and thus be present at low frequency in L. robustum and cannot be taken as evidence of introgression. In addition, three other bands were present only in introduced LRW and L. sinense. These three bands and the band shared between Rc21 and L. ovalifolium could result from introgression. However, Sri Lankan LRW shares two bands with L. sinense and one with L. ovalifolium, none of which was present in introduced material. As neither L. sinense nor L. ovalifolium occur in Sri Lanka, these three shared bands must either be symplesiomorphic in Ligustrum, or nonhomologous between introduced LRW and the other species. Therefore, either of these explanations might apply to the bands shared between introduced LRW and the other two species. Consequently, these shared bands do not constitute good evidence for introgression.
The material used in this study was collected as random samples from as broad a range as possible across La Réunion, to detect infraspecific variation and the possibility of multiple introductions. However, a future study concentrating on seeking introgression should sample extensively from LRW populations in close proximity of naturalised (or cultivated) material of one of the other species. The RAPD marker set generated here would provide a valuable tool for such a study.
Source area within Sri Lanka
AMOVA showed that there was no significant RAPD variation between LRW material sampled from different provinces in Sri Lanka (ie Uva, Central and Sabaragamuwa Provinces), nor between this material and introduced material from La Réunion. Consequently, it was not possible to pinpoint a part of the Sri Lankan population of LRW, which may have served as the source of introduced material.
The absence of significant variation of LRW between regions of Sri Lanka probably reflects extensive gene flow, perhaps due to long seed dispersal by birds, and also the fact that the provinces are not separated by natural boundaries. Given the low level of between-region variation, the exact source of introduction within Sri Lanka is probably not important for locating a biological control organism; a suitable organism would probably not discriminate between material throughout the range of LRW in Sri Lanka.
Conclusions
Both chloroplast DNA and RAPD data provide strong evidence that naturalised material of L. robustum in the Mascarene Islands of La Réunion and Mauritius is derived from Sri Lankan L. robustum ssp. walkeri. No evidence was found of introgression in the introduced material, although more detailed studies are required to rule out this possibility entirely. RAPDs indicate that L. robustum ssp. walkeri is more similar to L. perrottetii than it is to L. robustum ssp. robustum, and hence could be elevated to specific status or united with L. perrottetii as ‘L. perrottetii ssp. walkeri’. Based on this relationship, if a suitable biological control organism is not found in Sri Lanka, then the south Indian range of L. perrottetii would offer the next best chance of finding a useful control organism for invasive L. robustum walkeri in the Mascarene Islands.
References
Abbott RJ (1992). Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol Evol 7: 401–405.
Ansellem L, Noyer JL, Le Bourgeous T, Hossaert-McKey M (2000). Comparison of the invasive weed Rubus alceifolius Poir. (Rosaceae) in its native range and in areas of introduction, using Amplified fragment length polymorphisms (AFLP) markers. Mol Ecol 9: 443–455.
Daehler CC, Strong DR (1997). Hybridisation between introduced smooth cordgrass (Spartina alterniflora; Poaceae) and native California cordgrass (S. foliosa) in San Francisco Bay, California, USA. Am J Bot 84: 607–611.
Ellstrand NC, Schierenbeck K (2000). Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci USA 97: 7043–7050.
Excoffier L, Smouse PE, Quattro JM (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491.
Freischmann K (1997). Invasion of alien woody plants on the islands of Mahe and Silhouette. J Veg Sci 8: 5–12.
Green PS (1985). Ligustrum robustum subsp. walkeri. Kew Bull 40: 130.
Green PS (1987). Ligustrum, Oleaceae. In: Dassanayake MD, Fosberg FR (eds) A Revised Handbook to the Flora of Ceylon, Amerind publishing Co., New Delhi, India. Vol. 6, pp 270–271.
Green PS (1990). Ligustrum (Oleaceae) in southern India. Kew Bull 45: 693–696.
Haridasan K, Rao RR (1985). Forest Flora of Meghalaya Volume I: Ranunclulaceae to Cornaceae, Bishen Singh Mahendra Pal Singh: Dehra, India.
Hollingsworth ML, Hollingsworth PM, Jenkins GI, Bailey JP, Ferris C (1998). The use of molecular markers to study patterns of genotypic diversity in some invasive alien Fallopia spp. (Polygonaceae). Mol Ecol 7: 1681–1691.
Jansen RK, Palmer JD (1987). Chloroplast DNA from lettuce and Barnadesia (Asteraceae): structure, gene localisation and characterization of a large inversion. Curr Genet 11: 553–564.
Kuehn MM, Minor JE, White BN (1999). An examination of hybridization between the cattail species Typha latifolia and Typha angustifolia using random amplified polymorphic DNA and chloroplast DNA markers. Mol Ecol 8: 1981–1990.
Lavergne C, Rameau J-C, Figier J (1999). The invasive woody weed Ligustrum robustum subsp. walkeri threatens native forests on La Réunion. Biol Invasions 1: 377–392.
Macdonald IAW, Ortiz L, Lawesson JE, Nowak JB (1988). The invasion of highlands in Galapagos by the red Quinine-tree Cinchona succirubra. Environ Conser 15: 215–220.
Macdonald IAW, Thebaud C, Strahm WA, Strasberg D (1991). Effects of alien plant invasions on native vegetation remnants on La Réunion (Mascarene Islands, Indian Ocean). Environ Conser 18: 51–61.
Matthew KM (1999). Flora of the Palni Hills, South India, SCTP Offset Press: Christianpet, Vellore, India.
Meyer J-Y, Florence J (1996). Tahiti's native flora endangered by the invasion of Miconia calvescens DC. (Melastomataceae). J Biogeogr 23: 775–781.
Miller JT, Spooner DM (1996). Introgression of Solanum chacoense (Solanum sect Petota): upland populations reexamined. Syst Bot 21: 461–475.
Milne RI, Abbott RJ (2000). Origin and evolution of invasive naturalised material of Rhododendron ponticum L. in the British Isles. Mol Ecol 9: 541–556.
Milne RI, Abbott RJ, Wolff K, Chamberlain DF (1999). Hybridization among sympatric species of Rhododendron: (Ericaceae) in Turkey: morphological and molecular evidence. Am J Bot 86: 1776–1785.
Montaldo NH (1993). Avian dispersal and reproductive success of 2 species of Ligustrum (Oleaceae) in a subtropical forest relict in Argentina. Revista Chilena de Historia Natural 66: 75–85.
Mooney HA, Cleland ES (2001). The evolutionary impact of invasive species. Proc Natl Acad Sci USA 98: 5446–5451.
Novak SJ, Mack RN, Soltis PS (1993). Genetic variation in Bromus tectorum (Poaceae): introduction dynamics in North America. Can J Bot 71: 1441–1448.
Paterson IG, Snyder M (1999). Genetic evidence supporting the taxonomy of Geum peckii (Rosaceae) and G. radiatum as separate species. Rhodora 101: 325–340.
Perron M, Bousquet J (1997). Natural hybridization between black spruce and red spruce. Mol Ecol 6: 725–734.
Rieseberg LH, Wendel JF (1993). Introgression and its consequences in plants. In: Harrison R (ed) Hybrid Zones and the Evolutionary Process, Oxford University Press: Oxford. pp 70–109.
Rohlf FJ (1998). NTSYSpc: Numerical Taxonomy System, Version 2.02i, Exeter publishing Ltd, Setauket: New York.
Schneider S, Roessli D, Excoffier L (2000). Arlequin Version 2.001: A Software for Population Data Analysis, Genetics and Biometry Laboratory, University of Geneva: Switzerland.
Scott LJ, Graham GC, Hannan Jones M, Yeates DK (1997). DNA profiling resolves the limited importance of flower colour in defining varieties of Lantana camara. Electrophoresis 18: 1560–1653.
Smith CW (1989). Non-native plants. In: Stone CP, Stone DB (eds) Conservation Biology in Hawaii, Cooperative National Park Resource Studies Unit, University of Hawaii at Manoa, University of Hawaii Press: Honolulu. pp 60–68.
Spooner DM, Ugarte ML, Skroch PW (1997). Species boundaries and interrelationships of two closely related sympatric diploid wild potato species, Solanum astleyi and S. boliviense, based on RAPDs. Theor Appl Genet 95: 764–771.
Strasberg D (1995). Processus d'invasion par les plantes introduites a La Réunion et dynamique de la vegetation sur les coulees volcaniques. Ecologie 26: 169–180.
Strefeler MS, Darmo E, Becker RL, Katovich EJ (1996). Isozyme characterisation of genetic diversity in Minnesota populations of purple loosestrife Lythrum salicaria (Lythraceae). Am J Bot 83: 265–273.
Stromayer KAK, Warren RJ, Johnson AS, Hale PE, Rogers CL, Tucker CL (1998). Chinese privet and the feeding ecology of white-tailed deer: the role of an exotic plant. J Wildlife Manage 62: 1321–1329.
Swofford, DL (2002). PAUP*: phylogenetic analysis using parsimony *(and other methods). Version 4. Sinauer Associates, Sunderland, MA.
Vila M, D'Antonio CM (1998). Fitness of invasive Carpobrotus (Aizoaceae) hybrids in coastal California. Ecoscience 5: 191–199.
Williamson M (1996). Biological Invasions, Chapman & Hall: London.
Acknowledgements
Thanks are due to Richard Shaw and Christophe Lavergne for collection of material, and to two anonymous reviewers for comments on the manuscript. This research was financially supported by La Réunion district, through a contract with CAB International (Marché DEAT3/970354).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Milne, R., Abbott, R. Geographic origin and taxonomic status of the invasive Privet, Ligustrum robustum (Oleaceae), in the Mascarene Islands, determined by chloroplast DNA and RAPDs. Heredity 92, 78–87 (2004). https://doi.org/10.1038/sj.hdy.6800385
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800385
Keywords
This article is cited by
-
Genetic exchange between two freshwater apple snails, Pomacea canaliculata and Pomacea maculata invading East and Southeast Asia
Biological Invasions (2013)
-
Identification of new polymorphic regions and differentiation of cultivated olives (Olea europaea L.) through plastome sequence comparison
BMC Plant Biology (2010)
-
Hybridization and the evolution of invasiveness in plants and other organisms
Biological Invasions (2009)
-
On the origin of the invasive olives (Olea europaea L., Oleaceae)
Heredity (2007)