Abstract
We combined pedigree data with data derived from 14 microsatellite loci to investigate genetic diversity and its maintenance in the captive source population for the reintroduction of the bearded vulture into the Alps. We found the captive population to be genetically more variable than the largest natural population in Europe, both in terms of mean number of alleles per locus and mean observed and expected heterozygosity. Allelic diversity of the captive population was higher than, and mean heterozygosity measurements were comparable with the ones found in two large, extinct populations from Sardinia and the Alps represented by museum specimens. The amount of genetic variability recruited with the founders was still present in the captive population of the year 2000, mainly because the carriers of rare alleles were still alive. However, the decline in expected heterozygosity and the loss of alleles over generations in captivity was significant. Point estimates of effective population size, Ne, based on pedigree data and estimates of effective number of breeders, Nb, based on allele frequency changes, ranged from 20 to 30 and were significantly smaller than the census size. The results demonstrate that the amount of genetic variability in the captive bearded vulture population is comparable or even larger than the amount present in natural populations. However, the population is in danger to lose genetic variability over time because of genetic drift. Management strategies should therefore aim at preserving genetic variability by minimising kinship, and at increasing Ne by recruiting additional founders and enhancing gene flow between the released, the captive and natural populations.
Similar content being viewed by others
Introduction
The preservation of genetic variation is of special importance in captive breeding populations, which provide a reservoir of genetic material to be used to re-establish or reinforce populations in the wild (Lacy, 1993a). However, captive populations usually are derived from a small number of individuals and genetic variability may be lost not only during the founding event, but also during subsequent generations when the population remains small and exchange with other populations is minimal (Richards, 2000). The amount of genetic diversity lost over time is determined not simply by the number of individuals, but by the genetically effective population size, Ne (Wright, 1969). The number Ne is therefore an important factor in genetic conservation strategies to maximise genetic variability, as are the degrees of consanguinity, and the amount of genetic variation present in a population (Ballou and Foose, 1996). Assuming that the founders are unrelated and come from an initially outbred population, the genealogy of the individuals born in captivity allows an estimation of the change in average inbreeding coefficients over generations and thus the calculation of Ne (Blackwell et al, 1995). However, the basic assumption of unrelated founders may be incorrect, particularly given the often imprecise nature of information on their origin. In addition, pedigree data only help to estimate loss of genetic variability over time but can give no indication about the amount of genetic variability initially present among the founders.
In the present study, we therefore combine information from pedigree data and microsatellite DNA markers to estimate the amount and distribution of genetic variability present in the captive bearded vulture population. Over 100 young birds from this population have been released since 1986 to re-establish a bearded vulture population in the Alps. The original population of the Alps became completely extinct at the beginning of the last century, mainly because of hunting (Mingozzi and Estève, 1997). The captive population currently consists of about 120 birds kept in several zoos and breeding centres located in Europe, Asia and the United States. Its studbook lists 61 founders, of which 39 had reproduced by the year 2000. However, because the offspring of three founder birds died, all captive-born birds (F1 and F2 generations) descend from only 36 founder birds. Further, genetic similarity among the founder individuals can generate an effective population size (Ne) considerably smaller than the number of individuals that can be counted (Wright, 1969). In the captive bearded vulture population, information about the relatedness and geographic origin of the founders is incomplete. Many of them are known to come from several regions within the former Soviet Union, Afghanistan, the Pyrenees and Greece, but the origin of 14 individuals is unknown (Frey et al, 1995). Moreover, birds available for breeding vary in their reproductive success more strongly than in an assumed Poisson distribution of fecundity (Hartl and Clark, 1989). This may further reduce Ne in the captive bearded vulture population (Nunney, 1993).
Based on 14 microsatellite DNA markers, we determined the amount and distribution of genetic variability present in the captive bearded vulture (Gypaetus barbatus Linnaeus) population. We compared this variability with that of the largest extant bearded vulture population in Europe, the population from the Spanish Pyrenees, and with the variability that existed in two currently extinct large European populations from the last century, the one in the Alps and the one from Sardinia. In addition, we compare inbreeding coefficients and effective population size estimated using pedigree data with the estimates obtained from microsatellite data. Assuming neutrality of the analysed markers, discrepancies between pedigree- and microsatellite-based estimates will reflect consanguinity of the founder birds. The obtained information in this study will help to improve population-management efforts not only of the captive but, consequently, also of the reintroduced bearded vulture population and thus will help to reestablish successfully a self-sustaining population in the Alps.
Materials and methods
Modern samples
Blood samples in lysis buffer from 18 bearded vultures as well as already extracted DNA from nine bearded vultures from the wild population in the Spanish Pyrenees were kindly provided by Dr Juan Negro (Estacion Biologica, Doñana, Seville, Spain). Blood samples from 89 birds from the captive breeding population were collected by colleagues in the various zoos incorporated in the breeding programme and stored in lysis buffer (100 mM Tris-HCl, pH 8.0; 10 mM EDTA, pH 8.0; 10 mM NaCl; 0.5% SDS (w/v)). In addition, feather samples from 50 captive birds were collected, including feathers from 16 museum specimens. Feather samples were stored in the dark at room temperature. We isolated total genomic DNA from blood samples using standard proteinase K and organic solvent purification (Sambrook et al, 1989). DNA from feather samples was extracted using DNeasy® tissue kit (Qiagen) according to the manufacturer's instruction. Genetic diversity in the captive and the Pyrenean population was assessed using 14 microsatellite markers developed for the bearded vulture. The primer sequences, the conditions for the amplification of each locus, and the methods for allele detection were as described in Gautschi et al (2000).
Ancient samples
Feather samples from two extinct bearded vulture populations, the original population from the Alps (N=26), and the population from Sardinia (N=23) were collected in several Museums across Europe (see the Appendix). Using a sterile scalpel blade, the feather was cut from the skin leaving a small piece of skin attached to the basal section of the feather. Samples were stored in the dark at room temperature until further processing. DNA was extracted as described above for modern feather samples but with special lab precautions required for ancient DNA (aDNA) samples (Gautschi, 2001). Genetic diversity was assessed using the shorter and more variable microsatellite loci described in Gautschi et al (2000): BV2, BV5, BV6, BV11, BV12, BV13, BV14 and BV20. For the loci BV5, BV11, BV12, BV13 and BV14, we designed new primers that would amplify fragments of about 100–150 bp (Table 1), as in many of the ancient DNA samples only short fragments could be amplified. Microsatellite PCR amplification of aDNA samples was performed in a 11 μl volume containing 1 μl of extracted DNA, 5 μl of HotstarTaq® master mix (Qiagen), 4 μl double distilled water, and 0.5 μM each of forward and reverse primers. We used the following thermotreatment: 40–45 cycles with 95°C for 30 s, locus-specific annealing temperature (see Gautschi et al (2000) and Table 1) for 30 s, and 72°C for 30 s. In the initial cycle, a prolonged denaturation step (95°C for 15 min) was included and the last cycle was followed by an extra 8 min extension. Amplified products were detected as described in Gautschi et al (2000).
Assessment of genetic variability and estimation of inbreeding
In the captive bearded vulture population, we can distinguish among three distinct generation classes: founders, their descendants (F1) and their grand-offspring (F2). Further some cross-generation pairs were formed of F1 individuals with those of the founder generation. Their offspring belong to a generation referred to as F1/F2. Here we take two different approaches. First, we treat generations as discrete temporal units. Second, we considered two arbitrary generations, since bearded vultures are long-lived with strongly overlapping generations. The first generation includes only the founders. The second generation includes all animals still alive in 2000.
For each generation in the captive population as well as for the wild Pyrenean and the extinct Alp and Sardinian population, we assessed allele frequencies, the mean number of alleles per locus, mean observed heterozygosity, HO, and mean unbiased expected heterozygosity HE using the Biosys-1 software (Swofford and Selander, 1981). Departures from Hardy–Weinberg equilibrium (HWE) and Wright's inbreeding coefficient FIS were assessed using the program Fstat version 2.9 (Goudet, 1999), treating each generation of the captive population as a distinct subpopulation. Alleles were permuted within subpopulations (or generations) to test the significance of FIS. Differences in HO, HE, and mean number of alleles between the captive and the wild populations, among populations and among the generations within the captive population were tested by an analysis of variance (ANOVA) with locus as random and population (or generation) as fixed factor. Although the derived measures HO and HE theoretically do not have a normal distribution, the empirical distribution of the residuals did follow a normal distribution as assumed in ANOVA. The loss of expected heterozygosity and loss of alleles over the generations were estimated by regression analysis using the natural logarithm of HE or the total number of alleles (Briscoe et al, 1992). After fitting the generation we added the term locus.
Temporal genetic differences within populations should not be described with conventional contingency tests (Waples, 1989a). We therefore used FST for direct comparisons between temporal degrees of differentiation within the captive population (Jehle et al, 2001). Individual inbreeding coefficients from pedigree data were calculated with the studbook computer program (Zschokke, 2000).
Estimation of effective population size
We estimated effective population size based on genetic data using the temporal method (Waples, 1989b). This method measures the change in allele frequency (also referred to as F) over time and is based on the rationale that this change is a function of Ne and the time elapsed in generations (t). For short periods of time (ie. t<<2Ne) and under the assumption that the changes are due to drift, the following approximation can be made (Waples, 1991):
We estimated F as the change in microsatellite allele frequencies from one generation in the captive bearded vulture population to the next. For simplicity, we eliminated the offspring of cross-generation pairs (ie the F1/F2 ‘generation’) because they could not be assigned clearly to a generation. For the computation, we followed Nei and Tajima (1981),

where K is the number of segregating alleles, and xi and yi are the allele frequencies of the ith allele in the temporal samples (Nei and Tajima, 1981; Waples, 1989b). We combined the Fc values over loci (Kj alleles at the jth lcous) with the following formula (Nei and Tajima, 1981):

Ne can then be calculated as

with S0 and St being the number of individuals sampled at time 0 and t, respectively, and, Nobs being the size of the population subject to sampling at time zero (Waples, 1989b; Jehle et al, 2001). If the changes in allele frequencies from parent to offspring are used to calculate Fc, as was the case here, the result in equation (4) reflects the Ne -related parameter Nb, the effective number of breeders (Scribner et al, 1997). For t<3, estimates of Nobs greatly influence the accuracy of the Ne or Nb estimates. Here we used the number of founders recorded in the studbook, regardless of whether they have reproduced or have been sampled (eg 61) for all comparisons of allele frequency changes starting from the founder generation. In comparisons of allele frequencies changes from the F1 to the F2 generation, we used the total number of adult birds in the F1 generation as Nobs (eg 71). Confidence intervals for Nb were determined according to Waples (1989b). First, 1−α confidence limits to F were determined

which were then inserted into equation (4) to obtain confidence limits to Nb.
Calculations of Ne based on pedigree data involve the estimation of changes in inbreeding coefficient in a population over one (or more) generation. An estimate for Ne can then be obtained from the formula

where t is the time in generations elapsed, and ΔF is the change in inbreeding coefficient (Falconer and Mackay, 1996). We calculated the mean inbreeding coefficient for each generation in the captive population using Proc Inbreed of the SAS computer program (SAS Institute Inc., 1990). Proc Inbreed uses pedigree data to calculate an inbreeding coefficient F, from the covariance coefficient or coefficient of relation (Falconer and Mackay, 1996). The change in the mean inbreeding coefficient per generation ΔF (Falconer and Mackay, 1996) was calculated as

where Ft is the mean population inbreeding coefficient at generation t and Ft−1 represents the mean population inbreeding coefficient for the preceding generation. Again, we eliminated the offspring of cross-generation pairs (ie the F1/F2 ‘generation’) from the calculations.
Results
Assessment of genetic variability
The captive bearded vulture population was polymorphic at all 14 microsatellite loci investigated with a mean number of almost seven alleles per locus (Table 2). The natural population from the Pyrenees exhibited lower genetic variability both in terms of mean number of alleles per locus (F1,13=17.45, P<0.01) and genetic diversity as measured by mean observed and expected heterozygosity (F1,13=4.6, P=0.051, and F1,13=6.74, P<0.05, respectively) (Table 2).
The Pyrenean population also showed a lower percentage of polymorphic loci (Table 2), because of the two fixed loci BV7 and BV8. Based on the analysis of data from eight microsatellite loci that were also used for museum specimens, the genetic variability as measured by observed and expected heterozygosity in the captive population was similar to the one found in both extinct natural populations (Table 3).
However, diversity as measured by the mean number of alleles per locus was higher in the captive population than in the populations from the Alps and Sardinia (P=0.062 and P<0.01, respectively). No significant difference could be detected between the Pyrenean population and the extinct populations in the mean number of alleles per locus and the observed heterozygosity, and only the difference between the expected heterozygosity of the Pyrenean and the population from the Alps was substantial (P=0.055) (Table 3).
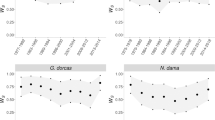
Genetic variability in the captive population was lost over the generations viewed as discrete units (Table 2), mainly because carriers of rare alleles have failed to pass them on. Figure 1 illustrates how several rare alleles present in the founder or F1 generation were not represented in the F1/F2 and F2 generation, and that the latter two generations were both fixed at locus BV8. The decline in the expected heterozygosity and the loss of alleles over generations was significant (P<0.01 for both) but not the decline in observed heterozygosity. When genetic variability present in the founders was compared with the one present in the breeding network of the year 2000, no significant difference could be detected in either of the genetic measurements.
Schematic illustration of relative allelic frequencies at 14 microsatellite loci for four different generations in the captive bearded vulture population; founders (black), F1 (dark grey), F1/F2 (light grey) and F2 (white). Circles represent distinct alleles, and their surface areas are directly proportional to their relative frequencies.
Temporal variation of allele frequencies, Hardy–Weinberg expectations and inbreeding
Figure 1 shows the allele frequencies for the respective microsatellite loci in the different generations of the captive population. Allele frequencies were subject to significant change between the founders and the F2 generation and between the F1 and the F1/F2 and the F1/F2 and the F2 generation (Table 4). No significant differentiation was found between the founders and the captive individuals present in the year 2000 (Table 4).
The observed genotype frequencies in the captive population, within the discrete generations in the captive populations, and the three natural population did not differ from Hardy–Weinberg expectations, not even at the locus level (data not shown). FIS values of the captive and the Pyrenean population ranged from −0.048 to 0.037. None of the values was significantly different from zero. The mean inbreeding coefficient (F), estimated from the pedigree doubled from the F1 to the F2 generation (mean FF1=0.0215 and mean FF2=0.0486, respectively) but only six individuals from the captive population have an individual inbreeding coefficient of greater than zero as revealed from pedigree data.
Estimations of effective number of breeders and effective population size
The value Fc, reflecting the variation in allele frequency changes, ranged between 0.030 and 0.047 (Table 5) and was larger when measured over longer time intervals (eg founder to F2). The effective number of breeders, Nb, calculated based on Fc, represented a fraction of the actual number of adults, Nobs, and none of the upper confidence limits reached Nobs. The point estimates of the effective number of breeders, Nb, ranged between 23 and 28 (Table 5). The ratio Nb/Nobs ranged between 0.32 and 0.46. Estimations of effective population size, Ne, based on the pedigree data, are summarised in Table 6. Both methods, the Ne estimation based on pedigree data and the Nb estimation based on microsatellite data, were about less than half the actual population size or the number of adults.
Discussion
Amount of genetic variability present in the captive and natural populations
Maintaining genetic diversity and demographic security are the primary population management goals for long-term conservation. The question of how much genetic diversity is required to sustain long-term fitness and evolutionary potential is, however, difficult to answer, as little is known about minimum genetic requirements of populations (Franklin and Frankham, 1998; Lynch and Lande, 1998). To determine the minimum genetic variability required in the captive bearded vulture population, we compared its genetic properties with the ones of the largest bearded vulture population in Europe, the population from the Spanish Pyrenees. We found the captive bearded vulture population to be genetically more variable than the Pyrenean population, both in terms of mean number of alleles per locus and mean observed and expected heterozygosity. The Pyrenean population, however, is geographically isolated and was reduced to only 50 pairs in the late 1970 (Hiraldo et al, 1979) and might therefore be genetically impoverished. In fact, a DNA fingerprinting study carried out on samples from this population revealed high band-sharing values (ie low level of genetic variation) compared to the ones found in other raptor species (Negro and Torres, 1999). The assumption of Negro and Torres (1999) of a genetic impoverishment of the Pyrenean population caused by a population bottleneck is supported by the higher genetic variability found in the two extinct populations from Sardinia and the Alps. Although the difference in genetic diversity between the Pyrenean population and the extinct Sardinian population was not significant, both allelic diversity and observed heterozygosity were substantially lower in Pyrenean population than in the population from the Alps. The captive population, on the other hand, revealed greater amounts of genetic variability, especially in terms of allelic diversity, than both extinct populations.
Loss of genetic variability and effective population size
Despite the diverse provenance of the founder birds in the captive bearded vulture population, no Wahlund effect (Hartl and Clark, 1989) could be detected. Neither the founders nor the subsequent generations within the captive population did significantly deviate from Hardy–Weinberg expectations. In addition, we found no indication of inbreeding as measured by the inbreeding coefficient FIS. So far this strategy of avoiding inbreeding by transfers of individuals among zoos to form breeding pairs seems to be successful (Frey et al, 1995). In addition, the genetic variability recruited from the founders was still present in the captive population in 2000. However, some rare alleles exist currently only in the founder generation and these may soon be lost if their carriers fail to pass them on. In fact, genetic variability in the discrete generations subsequent to the founders is reduced because of random genetic drift. Genetic drift, commonly the most powerful evolutionary force acting in small populations, results from random sampling of alleles at each generation. This can lead to changes in allele frequencies and fixation or loss of particular allelic variants. Allelic diversity is lost more rapidly than heterozygosity during a founding event (Allendorf, 1986) as we found here. Looking at discrete generations, we found no significant reduction in observed heterozygosity, although the F1 and F2 generations had significantly fewer alleles per locus on average. The rate of the loss of genetic diversity depends on the effective population size (Ne), with loss proceeding at a rate of 1/(2Ne) per generation (Hartl and Clark, 1989). Thus, minimising drift requires maintaining a large effective population size. Our Ne or Nb values for discrete generations were all around 20 or 30 depending on the method of estimation (pedigree or temporal, respectively). As we eliminated the offspring of the cross-generation pairs (ie F1/F2) from the calculations, our Ne estimates tend to be too small. However, only 6% of the living birds belong to the F1/F2 ‘generation’, and we therefore do not expect a large estimation bias. More important is the fact that bearded vultures are long-lived with strongly overlapping generations. The Ne estimates for the discrete generations will therefore not reflect the overall Ne, but rather give a point estimate for Ne in each generation. In each generation, we found an approximately two-fold discrepancy between the census size (Nobs) and the effective number of breeders or the effective population size. Effective population size is invariably lower than the census size, and estimates of Ne/N ratios in birds (with N being either census size or the number of sexually mature adults) range from 0.052 to 0.74 with a comprehensive mean of 0.32 (Frankham, 1995). The Ne/N ratio is predicted to be affected by factors such as unequal sex-ratio, variance in family sizes and fluctuations in population size over generations (Wright, 1969; Frankham, 1995). Relatedness among the founders would further reduce Ne (Wright, 1969) and could be detected by a difference between the Ne estimates from pedigree and data from genetic markers. This difference here is not significant. The discrepancy between Ne (or Nb) and Nobs detected in the captive bearded vulture population is most probably caused by unequal reproductive success of the breeding pairs (Frey et al, 1995) since the population exhibits neither an unequal sex ratio nor did we find evidence for closely related founder individuals.
Consequences for the management of the captive bearded vulture population
The amount of genetic variability in the captive bearded vulture population is comparable to or even larger than the amount present in large natural populations and so far, inbreeding could be avoided by transfers of individuals among zoos. However, the population is in danger to lose genetic variability over time because of genetic drift triggered by a small number of effective breeders. Clearly, management strategies should focus on preserving genetic diversity. The best approach to retain genetic diversity in captive populations has been shown to be minimising the overall level of relationship in the population, that is, minimising kinship (Montgomery et al, 1997). However, computer simulations using Vortex 8.41 (Lacy, 1993b) suggest that, even in the absence of inbreeding and catastrophic events, the captive population with the present size and age structure and a yearly release of eight birds over the next 30 years would lose more than 10% of its initial heterozygosity over the next 200 years (data not shown). Allelic diversity may even decrease more rapidly (Allendorf, 1986). In addition, the effective population size, required for a population to retain its evolutionary potential, may have to be as large as 500 or even 5000 (Franklin and Frankham, 1998). Apart from minimising kinship, management strategies in the bearded vulture reintroduction project should therefore focus on a rapid increase of the effective population size. This would not only reduce the potential negative effects of genetic drift, but would also buffer for demographic and environmental uncertainties (Menges, 1991) and perhaps allow the accumulation of new variation by protecting rare alleles from going extinct before they can spread (Ellstrand and Elam, 1993).
Avoiding variance in reproductive success of pairs by equalising family sizes will increase Ne and thus reduce the loss of genetic variability (Wright, 1969). However, equalisation of family size will change the selection regime, in particular for deleterious mutations (Couvet and Ronfort, 1994). Selection among the whole set of offspring produced will be replaced by selection among offspring produced in each family. The latter leads to a slower response of the character selected (Haldane, 1924). In some cases, for example, adaptation to captivity, a slower responde to selection is advantageous. In the case of deleterious mutations, however, it has been shown that the number of individual deaths necessary to eliminate lethal genes that are completely recessive or semidominant was larger under this selection regime, suggesting a higher mutation load (Campell, 1988). Thus, equalising family sizes to minimise the loss of genetic variability may lead to a higher frequency of deleterious mutations, and therefore to a loss of fitness (Couvet and Ronfort, 1994).
Apart from recruiting additional founder birds, the most promising approach for increasing effective population size is the strategy to enhance artificially gene flow between the captive and the released population. So far, birds are continuously released into the wild, which helps maintaining genetic variability in the introduced population that otherwise might experience excessive inbreeding. No immigration of birds from the released population back to the captive population is practised nor planned. However, immigration of birds from the released population could help reducing genetic drift, inbreeding and genetic adaptation to captivity in the captive population and both populations would benefit from a larger effective population size (Chesser, 1983). The amount of gene flow between the two populations should be high, because the reproductive variation within each population cannot be minimised easily by management strategies (Wang and Caballero, 1999). Clearly, it is unrealistic to assume that the captive and released population together will reach rapidly an effective population size of 5000 or even 500, needed to maintain evolutionary potential (Franklin and Frankham, 1998). However, gene flow (either natural or artificially aided) between the released population and natural populations in the Pyrenees, on Corsica, and in the Balkans could further enhance Ne and thus could help maintain the evolutionary potential of the bearded vulture across Europe and restoring genetic variation within small and endangered natural populations (Ingvarsson, 2001).
References
Allendorf FW (1986). Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol 5: 181–190.
Ballou JD, Foose TJ (1996). Demographic and genetic management of captive populations. In: Kleinman DG, Allen ME, Thompson KV, Lumpkin S (eds) Wild Mammals in Captivity, The University of Chicago Press: Chicago. pp 263–283.
Blackwell BF, Doerr PD, Reed JM, Walters JR (1995). Inbreeding rate and effective population size: a comparison of estimates from pedigree analysis and a demographic model. Biol Conserv 71: 299–304.
Briscoe DA, Malpica JM, Robertson A, Smith GJ, Frankham R, Banks RG et al (1992). Rapid loss of genetic variation in large captive populations of Drosophila flies: implications for the genetic management of captive populations. Conserv Biol 6: 416–425.
Campell RB (1988). Mating structure and the cost of deleterious mutation: postponing inbreeding. J Hered 79: 179–183.
Chesser RK (1983). Isolation by distance: relationship to the management of genetic resources. In: Schonewald-Cox CM, Chabers SM, MacBryde B, Thomas WL (eds) Genetics and Conservation: a Reference for Managing Wild Animal and Plant Populations, Benjamin/Cummings: Menlo Park, CA. pp 66–77.
Couvet D, Ronfort J (1994). Mutation load depending on variance in reproductive success and mating system. In: Loeschcke V, Tomiuk J, Jain SK (eds) Conservation Genetics, Birkhäuser: Basel. pp 55–68.
Ellstrand NC, Elam DR (1993). Population genetic consequences of small population-size – implications for plant conservation. Annu Rev Ecol Syst 24: 217–242.
Falconer DS, Mackay TFC (1996). Introduction to Quantitative Genetics, 4th edn. Longman: Essex.
Frankham R (1995). Effective population size/adult population size ratios in wildlife: a review. Genetet Res 66: 95–107.
Franklin IR, Frankham R (1998). How large must populations be to retain evolutionary potential. Anim Conserv 1: 69–73.
Frey H, Knotzinger O, Llopis Dell A (1995). The breeding network – an analysis of the period 1978 to 1995. In: Frey H, Kurzweil J, Bijleveld M (eds) Bearded Vulture Reintroduction into the Alps, Annual Report 1995, Foundation of the Conservation of the Bearded vulture: Wien. pp 13–38.
Gautschi B (2001). Conservation genetics of the bearded vulture (Gypaetus barbatus). PhD thesis, University of Zurich, Switzerland.
Gautschi B, Tenzer I, Müller JP, Schmid B (2000). Isolation and characterization of microsatellite loci in the bearded vulture (Gypaetus barbatus) and cross-amplification in three Old World vulture species. Mol Ecol 9: 2193–2195.
Goudet J (1999). FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9). Available from http://www.unil.ch/izea/softwares/fstat.html. updated from Goudet (1995).
Haldane JBS (1924). A mathematical theory of natural and artificial selection. Trans Camb Philos Soc 23: 19–41.
Hartl DL, Clark AG (1989). Principals of Population Genetics, 2nd edn. Sinauer Associates, Inc.: Sunderland, MA.
Hiraldo R, Elibes M, Calderon J (1979). El Quebrantahuesos Gypaetus barbatus L. Monografia 22, ICONA: Madrid.
Ingvarsson PK (2001). Restoration of genetic variation lost – the genetic rescue hypothesis. Tree 16: 62–63.
Jehle R, Arntzen WJ, Burke T, Krupa AP, Hödl W (2001). The annual number of breeding adults and the effective population size of syntopic newts (Triturus cristatus, T. marmoratus). Mol Ecol 10: 839–850.
Lacy RC (1993a). Impacts of inbreeding in natural and captive populations of vertebrates: implications for conservation. Perspect Biol Med 36: 480–496.
Lacy RC (1993b). VORTEX: a computer simulation model for population viability analysis. Wildl Res 20: 45–65.
Lynch M, Lande R (1998). The critical effective size for a genetically secure population. Anim Conserv 1: 70–72.
Menges ES (1991). The application of minimum viable population theory to plants. In: Falk DA, Holsinger KE (eds) Genetics and Conservation of Rare Plants, Oxford University Press: Oxford, UK. pp 45–61.
Mingozzi T, Estève R (1997). Analysis of a historical extirpation of the bearded vulture Gypaetus barbatus (L.) in the Western Alps (France–Italy): former distribution and causes of extirpation. Biol Conserv 79: 155–171.
Montgomery ME, Ballou JD, Nurthen RK, England PR, Briscoe DA, Frankham R (1997). Minimizing kinship in captive breeding programs. Zoo Biol 16: 377–389.
Negro JJ, Torres MJ (1999). Genetic variability and differentiation of two bearded vulture Gypaetus barbatus populations and implications for reintroduction projects. Biol Conserv 87: 249–254.
Nei M, Tajima F (1981). Genetic drift and estimation of effective population size. Genetics 98: 625–640.
Nunney L (1993). The influence of mating system and overlapping generations of effective population size. Evolution 47: 1329–1341.
Richards CM (2000). Inbreeding depression and genetic rescue in a plant metapopulation. Am Nat 155: 383–394.
Sambrook J, Fritsch EF, Maniatis T (1989). Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbour Laboratory: Cold Spring Harbour, NY.
SAS Institute Inc. (1990). SAS Language: Reference, First Edition Version 6 edit. SAS Institute Inc.: Cary, NC.
Scribner KT, Arntzen JW, Burke T (1997). Effective number of breeding adults in Bufo bufo estimated from age-specific variation at minisatellite loci. Mol Ecol 6: 701–712.
Swofford DL, Selander RB (1981). BIOSYS-1: a FORTRAN program for the comprehensive analysis of electrophoretic data in population genetics and systematics. J Hered 72: 281–283.
Wang J, Caballero A (1999). Developments in predicting the effective size of subdivided populations. Heredity 82: 212–226.
Waples RS (1989a). Temporal variation in allele frequencies: testing the right hypothesis. Evolution 43: 1236–1251.
Waples RS (1989b). A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics 121: 379–391.
Waples RS (1991). Genetic methods for estimating the effective size of cetacean populations. In: Hoelzel AR (ed) Genetic Ecology of Whales and Dolphins, Special Issue 13. The International Whaling Commission: Cambridge. pp 279–300.
Wright S (1969). Evolution and the Genetics of Populations: The Theory of Gene Frequencies. University of Chicago Press: Chicago. Vol. 2.
Zschokke S (2000). Studbook v. 3.2 for Macintosh. University of Basel, Basel.
Acknowledgements
We thank Dr Hans Frey (Veterinärmedizinisches Institut, Universität Wien, Austria) and Dr Juan Negro (Estación Biológica Doñana, Seville, Spain) for providing blood samples from the captive and the Pyrenean population. Thanks to all colleagues at various zoos and breeding centres for sending blood and feather samples. Special thanks goes to the Natural History Museums (Appendix) for permitting sample collection from their bearded vulture specimen. This work was supported by the Swiss National Science Foundation grant 31-49477.96 to JPM, BG and BS.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Summary of bearded vulture specimen sampled in 17 European Natural History Museums is presented in Table 7. Listed are the sample ID provided by the museums, the location of the museum (city and country), the provenance (location and country) of the sample, the year of collection and the population they were grouped to.
Rights and permissions
About this article
Cite this article
Gautschi, B., Müller, J., Schmid, B. et al. Effective number of breeders and maintenance of genetic diversity in the captive bearded vulture population. Heredity 91, 9–16 (2003). https://doi.org/10.1038/sj.hdy.6800278
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800278
Keywords
This article is cited by
-
Ex situ versus in situ Eurasian lynx populations: implications for successful breeding and genetic rescue
Conservation Genetics (2023)
-
Assessing evolutionary processes over time in a conservation breeding program: a combined approach using molecular data, simulations and pedigree analysis
Biodiversity and Conservation (2021)
-
Studbook and molecular analyses for the endangered black-lion-tamarin; an integrative approach for assessing genetic diversity and driving management in captivity
Scientific Reports (2020)
-
Low genetic diversity and shallow population structure in the endangered vulture, Gyps coprotheres
Scientific Reports (2019)
-
Microsatellite polymorphism in the endangered snail kite reveals a panmictic, low diversity population
Conservation Genetics (2018)