Abstract
The chromosomal localization of ribosomal DNA, and a 180 bp satellite DNA isolated from Spanish Eyprepocnemis plorans specimens, has been analysed in five Eyprepocnemidinae species collected in Russia and Central Asia. Caucasian E. plorans individuals carried each of the two DNAs, but the rDNA was limited to only two chromosomes (S9 and S11) in sharp contrast to Spanish specimens that show 4–8 rDNA clusters and to Moroccan specimens which carry rDNA in almost all chromosomes. The four remaining species, however, lacked the 180 bp tandem repeat, and showed rDNA clusters in one (S9 in Thisoicetrinus pterostichus), two (S9 and S10 in Eyprepocnemis unicolor; M8 and S11 in Heteracris adspersa), or three (S9, S10, and S11 in Shirakiacris shirakii) chromosome pairs. The implications of these findings for the evolution of these two chromosome markers in this group of species are discussed.
Similar content being viewed by others
Introduction
As the genome era progresses, it is being more apparent that eukaryote genomes contain a significant proportion of repetitive DNA of various types. Two of the tandem repetitive DNA classes, that is, ribosomal (rDNA) and satellite DNA (satDNA), are very different in genetic role (rDNA is very important functionally, but satDNA seems to be nonfunctional) but they share some evolutionary properties (eg concerted evolution).
Ribosomal DNA is arranged in tandem arrays (containing transcriptional units coding for three of the four rRNA types) located at one or more chromosome regions, the so-called nucleolus organizer regions (NORs). NORs are evolutionarily dynamic, which makes them excellent markers for phylogenetic studies (Cabrero and Camacho, 1986; Amemiya and Gold, 1988; Santos and Fox, 1988). Interspecies comparison of NOR number and chromosome location may be a good tool for macroevolutionary studies. Intraspecific variation, moreover, when detected, is indicative of the complex microevolutionary patterns shown by rDNA. The most remarkable conclusion of rDNA research at these two levels is that NORs seem to be able to spread through the genome thus creating new rDNA loci (Castro et al, 2001). Several spreading mechanisms have been suggested, for example, transposition (Schubert and Wobus, 1985), insertion of extrachromosomal rDNA amplified during oogenesis (Phillips et al, 1988), the presence of repetitive elements facilitating nonhomologous chromosome exchange (Maggini et al, 1991), and the amplification of minor rDNA loci (Dubcovsky and Dvörak, 1995).
The presence of satDNA is characteristic of most eukaryote organisms. It is restricted to heterochromatic chromosome regions and is typically noncoding. It usually consists of a tandemly repeated unit of size varying, in most cases, from 10 to 100 bp (Charlesworth et al, 1994).
SatDNA constitutes a rapidly evolving marker, which makes it useful for phylogenetic studies. Its presence or absence, and the divergence of sequences shared among species, may be powerful tools, throwing light on the evolutionary relationships among closely related species. Such information has proven to be useful in a variety of organisms, for example, insects (Bachmann and Sperlich, 1993; Juan et al, 1993; Bachmann et al, 1994; Pons et al, 1997), fishes (Garrido-Ramos et al, 1995; de la Herrán et al, 2001; Lanfredi et al, 2001), birds (Madsen et al, 1992), mammals (Hamilton et al, 1992; Wijers et al, 1993; Volobouev et al, 1995; Lee et al, 1999), and plants (Galasso et al, 2001).
The grasshopper Eyprepocnemis plorans harbours a B chromosome polymorphism which has become a paradigm of the long-term evolution of parasitic B chromosomes, since it has provided clear evidence for an arms race between the coevolving A and B chromosomes (Camacho et al, 1997). These B chromosomes are mainly made up of two repetitive DNAs, that is rDNA and a 180 bp satDNA, which are also present in many A chromosomes (López-León et al, 1994,1995b; Cabrero et al, 1999). To ascertain the origin of these B chromosomes is, therefore, necessary to investigate the presence and chromosome distribution of these two repetitive DNAs in geographically distant E. plorans populations as well as in some other closely related species, since such a study might uncover the ancestral patterns and the evolutionary pathways that could explain the present chromosome distribution. In this paper, we analyse the presence and chromosome localization of these two repetitive DNAs in five Eyprepocnemidine species from Russia and Central Asia, including E. plorans.
Materials and methods
A total of 24 adult male grasshoppers belonging to five Eyprepocnemidinae species, collected at several localities in Russia and Central Asia (see locations in Table 1 in Bugrov et al, 1999), were analysed by fluorescence in situ hybridization (FISH) and silver impregnation. Two of the species analysed belonged to the genus Eyprepocnemis (E. plorans, five males collected at Daghestan, North Caucasus, and E. unicolor, six males collected at Tajikistan), the three other species being Heteracris adspersa (five males from Daghestan), Thisoicetrinus pterostichus (five males from Daghestan), and Shirakiacris shirakii (three males from Primorskij kray).
Chromosome preparations were made by squashing two testis follicles in 50% acetic acid. After 10 min, the coverslip was removed with a razor blade after freezing the preparation by immersion in liquid nitrogen for a few seconds. Silver impregnation was performed following the technique described by Rufas et al (1982).
For FISH, in order to eliminate cell cytoplasm and thus facilitate probe accessibility, each slide was incubated in 150 μl of pepsin (50 μg/ml in 0.01N HCl) at 37°C in a humid chamber for 10–30 min. After three washes in distilled water, the slides were dehydrated in an ethanol series at 70, 90, and 100% for 3, 3, and 5 min, respectively, and then were air dried. Slides were stored at 60°C overnight before in situ hybridization.
FISH was performed with two different DNA probes: pTa71, which contains a 9-kb EcoRI repeat unit of rDNA isolated from Triticum aestivum (Gerlach and Bedbrook, 1979), and pEpD15, which contains a 180 pb DraI fragment of a tandemly repetitive DNA isolated from E. plorans (López-León et al, 1994,1995b). The probe DNA was labelled by nick translation with Fluorogreen 11-dUTP or Fluorored 11-dUTP, using standard techniques. FISH was performed following the technique described in López-León et al (1994). In brief, the two DNA probes were mixed to a final concentration of 5 ng/μl in a solution containing 40% formamide, 10% dextran sulfate, 0.1% sodium dodecyl sulphate (SDS) in 1 × SSC, and 70 ng/μl salmon sperm. Chromosomal DNA was denaturized by incubation with the hybridization mixture (30 μl) in a hot plate at 80°C for 8 min. Hybridization was performed at 37°C overnight. After two washes in 2 × SSC at 37°C, one in 2 × SSC at room temperature and one in 4 × SSC/Tween 20 (5 min. each), slides were counterstained with 2 μg/ml of DAPI (4′6 diamidino-2-phenylindole) and mounted in antifade solution (Vectashield H-1000).
Photographs were taken on Fujichrome 400 Provia colour film. Slides were digitized with a Hewlett-Packard Photo Smart scanner and the figures were composed with Adobe Photoshop and Microsoft Word.
Results
As described elsewhere (Bugrov et al, 1999), the five species show 2n=22+X0♂/XX♀ acrocentric chromosomes, with two long (L1 and L2), six medium (M3–M8) and three short (S9–S11) autosomes, and the X chromosome showing a size similar to that of the longest M autosomes.
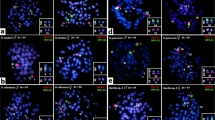
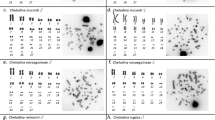
Caucasian males of E. plorans show two rDNA clusters located on the S9 and S11 autosomes (Figure 1a). The 180 bp tandem DNA repeat, however, is located on paracentromeric regions of the X chromosome and eight autosomal bivalents (excluding M8, S9, and S10) (Figure 1a,b). Note that S11 is the only standard chromosome carrying the two repetitive DNAs. None of the four other Eyprepocnemidinae species carried the E. plorans repetitive DNA, suggesting that it is species specific. In consistency with rDNA location, silver impregnation showed active NORs only in bivalents S9 and S11 (Figure 2a).
In the remaining species, the rDNA was located on a variable number of bivalents: one in Thisoicetrinus pterostichus (S9) (Figure 1e), two in Eyprepocnemis unicolor (S9 and S10) (Figure 1c) and Heteracris adspersa (M8 and S11) (data not shown), and three in Shirakiacris shirakii (S9, S10, and S11) (Figure 1d). Silver impregnation showed that all rDNA clusters were active in all four species (Figure 2; data not shown for T. pterostichus).
Discussion
Species-specific satDNA has been found in some cases, for example, the cave cricket Dolichopoda schiavazzii (Bachmann et al, 1994). In these cases, this marker provides very useful information for microevolutionary studies. When the same satDNA is shared by other species, however, it becomes a valuable marker for phylogenetic studies. For example, it may be shared by part of a genus (eg in Tribolium flour-beetles, Juan et al, 1993), a complete genus (eg Peromyscus rodents, Hamilton et al, 1992, Pimelia beetles, Pons et al, 1997, and Lens legumes, Galasso et al, 2001), several genera in a same family (eg the RBMII sequences in the avian Anatidae family, Madsen et al, 1992, and the DraI satDNA in the Sparidae fish family, de la Herrán et al, 2001), and even a whole family (eg the EcoRI satDNA in the Sparidae fish family, de la Herrán et al, 2001) or order (eg the human gamma-X centromeric satellite DNA, which seems to be widespread among primates, Lee et al, 1999).
The 180 bp tandem DNA repeat isolated from Spanish specimens of E. plorans (López-León et al, 1995b) is present in Caucasian males of this species, but not in the four remaining Eyprepocnemidinae species analysed. It was absent from E. unicolor suggesting that this satDNA is specific for E. plorans and is not a characteristic of the genus. Chromosomal location of this satDNA, in paracentromeric regions of most chromosomes, is roughly similar in E. plorans populations from such distant places as Spain (López-León et al, 1994), Morocco (Bakkali, 2001), and the Caucasus (this paper). In a recent analysis of 12 Spanish populations, we have found the presence of this satDNA in seven to 10 out of the 12 chromosomes of E. plorans, with the S10 chromosome lacking satDNA in all populations (as in our present study) and the S9 carrying it only in populations from southern Spain (Cabrero et al, 2003). This interpopulational resemblance suggests that this satDNA appeared soon in the evolution of E. plorans, most likely in the ancestral African populations before the colonization of the remaining regions, since Eyprepocnemis is a genus of African origin (Dirsh, 1958). It would be, however, interesting, to compare the DNA sequence of this repeat among specimens from the three geographic regions. In addition, the analysis of presence, chromosome localization, and sequence in the four E. plorans subspecies that have been described in Africa (Dirsh, 1958) could be very informative for the evolution of this species.
The rDNA is restricted to one to three of the smallest chromosomes (M8–S11) in the five Eyprepocnemidinae species analysed here. This suggests that the ancestral number of rDNA clusters in the Eyprepocnemidinae is restricted to one to three of the four smallest autosomes (M8–S11). In some animal groups, for example, fishes, the existence of a single rDNA cluster is considered a plesiomorphic character (see Jankun et al, 2001), although many examples of multichromosomal location of rDNA have been described (see Castro et al, 2001). In the Eyprepocnemidinae grasshoppers analysed here, the most conserved rDNA cluster is that located on the S9 chromosome, which is present in four of the five species analysed. This chromosome constitutes the so-called ‘megameric’ bivalent during male meiosis, which shows an allocyclic behaviour similar to the X univalent, with coincident changes in heteropycnosis (Hewitt, 1979). In a study of chromosome NOR location in 21 species of grasshoppers, by means of silver impregnation, Rufas et al (1985) suggested the megameric chromosome to be one of the ancestral NOR locations within the family Acrididae. It is thus possible that the monochromosomal ancestral location of rDNA in Eyprepocnemidinae grasshoppers was the megameric autosome pair, a condition that is still conserved in Thisoicetrinus pterostichus. In the remaining species, the number of rDNA clusters has increased by the presence of additional rDNA clusters in one (S11 in E. plorans and S10 E. unicolor) or two small chromosomes (S10 and S11 in Shirakiacris shirakii). In Heteracris adspersa, however, the ancestral S9 cluster has been lost and the rDNA is located on M8 and S11 chromosomes.
There is no clear explanation for interspecies differences in the number of rDNA clusters in the genome. Some authors have explored the possibility of a relation with anatomical differences, but the number of rDNA sites does not show any consistency with morphologically based phylogeny in carabid beetles (Sánchez-Gea et al, 2000). Other authors have found a weak but positive correlation between chromosome number and the number of rDNA loci (Hirai et al, 1994). But this is not applicable to the Eyprepocnemidinae grasshoppers analysed here, because the observed variation in the number of rDNA clusters was found in species with the same chromosome number.
The presence of rDNA only in the S9 and S11 chromosomes of Caucasian specimens of E. plorans is in sharp contrast with its presence in most chromosomes of Spanish and Moroccan specimens (López-León et al, 1994; Bakkali et al, 2001; Cabrero et al, 2003). This shows the occurrence of a dramatic spread of rDNA over the whole genome in western populations of E. plorans. Especially remarkable is the absence of rDNA from the X chromosome in Caucasian E. plorans and the four other Eyprepocnemidinae species analysed, a chromosomal location which constitutes one of the principal active NORs in western individuals (López-León et al, 1995a; Cabrero et al, 1987; Bakkali et al, 2001) which suggests that this absence is ancestral.
References
Amemiya CT, Gold JR (1988). Chromosomal NORs as taxonomic and systematic characters in North American cyprinid fishes. Genetica 76: 81–90.
Bachmann L, Sperlich D (1993). Gradual evolution of a specific satellite DNA family in Drosophila ambigua, D. tristis, and D. obscura. Mol Biol Evol 10: 647–659.
Bachmann L, Venanzetti F, Sbordoni V (1994). Characterization of a species-specific satellite DNA family of Dolichopoda schiavazzii (Orthoptera, Raphidophoridae) cave crickets. J Mol Evol 39: 274–281.
Bakkali M (2001). Evolución de los cromosomas B del saltamontes Eyprepocnemis plorans en Marruecos. PhD Thesis, Universidad de Granada, Spain.
Bakkali M, Cabrero J, López-León MD, Perfectti F, Camacho JPM (2001). Population differences in the expression of nucleolus organizer regions in the grasshopper Eyprepocnemis plorans. Protoplasma 217: 185–190.
Bugrov A, Warchałowska-Śliwa E, Vysotskaya L (1999). Karyotypic features of Eyprepocnemidinae grasshoppers from Russia and Central Asia with reference to the B chromosomes in Eyprepocnemis plorans (Charp.). Folia Biologica (Kraków) 47: 97–104.
Cabrero J, Alché JD, Camacho JPM (1987). Effects of B chromosomes of the grasshopper Eyprepocnemis plorans on nucleolar organiser regions activity. Activation of a latent NOR on a B chromosome fused to an autosome. Genome 29: 116–121.
Cabrero J, Camacho JPM (1986). Cytogenetic studies in gomphocerine grasshoppers. II. Chromosomal location of active nucleolar organizing regions. Can J Genet Cytol 28: 540–544.
Cabrero J, López-León MD, Bakkali M, Camacho JPM (1999). Common origin of B chromosome variants in the grasshopper Eyprepocnemis plorans. Heredity 83: 435–439.
Cabrero J, Perfectti F, Gómez R, Camacho JPM, López-León MD (2003). Population variation in the A chromosome distribution of satellite DNA and ribosomal DNA in the grasshopper Eyprepocnemis plorans. Chromosome Res. Chromosome Res. (in press).
Camacho JPM, Shaw MW, López–León MD, Pardo MC, Cabrero J (1997). Population dynamics of a selfish B chromosome neutralized by the standard genome in the grasshopper Eyprepocnemis plorans. Am Nat 149: 1030–1050.
Castro J, Rodríguez S, Pardo BG, Sánchez L, Martínez P (2001). Population análisis of an unusual NOR-site polymorphism in brown trout (Salmo trutta L.). Heredity 86: 291–302.
Charlesworth B, Sniegowski P, Stephan W (1994). The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220.
De la Herrán R, Ruiz Rejón C, Ruiz Rejón M, Garrido-Ramos MA (2001). The molecular phylogeny of the Sparidae (Pisces, Perciformes) based on two satellite DNA families. Heredity 87: 691–697.
Dirsh VM (1958). Revision of the genus Eyprepocnemis Fieber, 1853 (Orthoptera: Acridoidea). Proc R Entomol Soc London B 27: 33–45.
Dubcovsky J, Dvörak J (1995). Ribosomal RNA multigene loci: nomads of the Triticeae genomes. Genetics 140: 1367–1377.
Galasso I, Schmidt T, Pignone D (2001). Identification of Lens culinaris ssp. culinaris chromosomes by physical mapping of repetitive DNA sequences. Chromosome Res 9: 199–209.
Garrido-Ramos MA, Jamilena M, Lozano R, Cárdenas S, Ruiz Rejón C, Ruiz Rejón M (1995). Phylogenetic relationships of the Sparidae family (Pisces, Perciformes) inferred from satellite-DNA. Hereditas 122: 1–6.
Gerlach WL, Bedbrook JR (1979). Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7: 1869–1885.
Hamilton MJ, Hong G, Wichman HA (1992). Intragenomic movement and concerted evolution of satellite DNA in Peromyscus: evidence from in situ hybridization. Cytogenet Cell Genet 60: 40–44.
Hewitt GM (1979). Grasshoppers and Crickets. Animal Cytogenetics 3, Insecta 1, Orthoptera. Gebrüder Borntraeger: Berlin-Stuttgart.
Hirai H, Yamamoto M-T, Ogura K, Satta Y, Yamada M, Taylor RW, Imai HT (1994). Multiplication of 28S rDNA and NOR activity in chromosome evolution among ants of the Myrmecia pilosula species complex. Chromosoma 103: 171–178.
Jankun M, Martínez P, Pardo BG, Kirtiklis L, Rab P, Rabova M, Sánchez L (2001). Ribosomal genes in Coregonid fishes (Coregonus lavaretus, C. albula and C. peled) (Salmonidae): single and multiple nucleolus organizer regions. Heredity 87: 672–679.
Juan C, Vazquez P, Rubio JM, Petitpierre E, Hewitt GM (1993). Presence of highly repetitive DNA sequences in Tribolium flour-beetles. Heredity 70: 1–8.
Lanfredi M, Congiu L, Garrido-Ramos MA, de la Herrán R, Leis M, Chicca M, Rossi R, Tagliavini J, Ruiz Rejón C, Ruiz Rejón M, Fontana F (2001). Chromosomal location and evolution of a satellite DNA family in seven sturgeon species. Chromosome Res 9: 47–52.
Lee C, Stanyon R, Lin C-C, Ferguson-Smith MA (1999). Conservation of a human gamma-X centromeric satellite DNA among primates with an autosomal localization in certain Old World monkeys. Chromosome Res 7: 43–47.
López-León MD, Cabrero J, Camacho JPM (1995a). Changes in NOR activity pattern in presence of supernumerary heterochromatin in the grasshopper Eyprepocnemis plorans. Genome 38: 68–74.
López-León MD, Neves N, Schwarzacher T, Heslop-Harrison TS, Hewitt GM, Camacho JPM (1994). Possible origin of a B chromosome deduced from its DNA composition using double FISH technique. Chromosome Res 2: 87–92.
López-León MD, Vázquez P, Hewitt GM, Camacho JPM (1995b). Cloning and sequence analysis of an extremely homogeneous tandemly repeated DNA in the grasshopper Eyprepocnemis plorans. Heredity 75: 370–375.
Madsen C, McHugh KP, de Kloet SR (1992). Characterization of a major tandemly repeated DNA sequence (RBMII) prevalent among many species of waterfowl (Anatidae). Genome 35: 1037–1044.
Maggini FT, Cremonini R, Zolfino C, Tucci GF, D'Ovidio R, Delre V, DePace C, Scarascia Mugnozza GT, Cionini PG (1991). Structure and chromosomal localization of DNA sequences related to ribosomal subrepeats in Vicia faba. Chromosoma 100: 229–234.
Phillips RB, Pleyte KA, Hartley SE (1988). Stock-specific differences in the number and chromosome positions of the nucleolar organizer regions in Arctic charr (Salvelinus alpinus). Cytogenet Cell Genet 48: 9–12.
Pons J, Bruvo B, Juan C, Petitpierre E, Plohl M, Ugarkovic D (1997). Conservation of a satellite DNA in species of the genus Pimelia (Tenebrionidae, Coleoptera). Gene 205: 183–190.
Rufas JS, Esponda P, Gosálvez J (1985). NOR and nucleolus in the spermatogenesis of acridoid grasshoppers. Genetica 66: 139–144.
Rufas JS, Iturra P, De Souza W, Esponda P (1982). Simple silver staining procedure for the localization of nucleolus and nucleolar organizer under light and electron microscopy. Arch Biol 93: 267–274.
Sánchez-Gea JF, Serrano J, Galián J (2000). Variability in rDNA loci in Iberian species of the genus Zabrus (Coleoptera: Carabidae) detected by fluorescence in situ hybridization. Genome 43: 22–28.
Santos JL, Fox DP (1988). A study of nucleolus organiser regions (NORs) in the subfamily gomphocerinae (Acrididae; Orthoptera) by means of an acridine orange staining procedure. Genet (Life Sci Adv) 7: 27–32.
Schubert I, Wobus U (1985). In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92: 143–148.
Volobouev V, Vogt N, Viegas-Péquignot E, Malfoy B, Dutrillaux B (1995). Characterization and chromosomal location of two repeated DNAs in three Gerbillus species. Chromosoma 104: 252–259.
Wijers E, Zijlstra C, Lenstra JA (1993). Rapid evolution of horse satellite DNA. Genomics 18: 113–117.
Acknowledgements
This study was supported by grants from the Spanish Ministerio de Ciencia y Tecnología (BOS2000-1521) and Plan Andaluz de Investigación, Grupo no. CVI-165.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cabrero, J., Bugrov, A., Warchałowska-Śliwa, E. et al. Comparative FISH analysis in five species of Eyprepocnemidine grasshoppers. Heredity 90, 377–381 (2003). https://doi.org/10.1038/sj.hdy.6800255
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800255
Keywords
This article is cited by
-
Modified Flame Drying Technique to Study Chromosomes of Grasshopper’s Testis
Proceedings of the Zoological Society (2015)
-
Disparate molecular evolution of two types of repetitive DNAs in the genome of the grasshopper Eyprepocnemis plorans
Heredity (2014)
-
HP1 knockdown is associated with abnormal condensation of almost all chromatin types in a grasshopper (Eyprepocnemis plorans)
Chromosome Research (2014)
-
Ribosomal DNA is active in different B chromosome variants of the grasshopper Eyprepocnemis plorans
Genetica (2013)
-
Gypsy, RTE and Mariner transposable elements populate Eyprepocnemis plorans genome
Genetica (2012)