Abstract
We describe a young woman with Prader–Willi syndrome (PWS) due to a mosaic imprinting defect. Three independent assays revealed a reduced proportion of nonmethylated SNURF-SNRPN alleles in peripheral blood DNA: methylation-specific PCR followed by denaturing high-performance liquid chromatography (MSP/DHPLC), methylation-sensitive restriction enzyme analysis and methylation-specific real-time PCR analysis. Microsatellite analysis and fluorescence in situ hybridisation revealed apparently normal chromosomes 15 of biparental origin. Based on the MSP/DHPLC and real-time PCR results, we estimate that approximately 50% of the patient's blood cells have an imprinting defect and 50% of the cells are normal. Apart from a rather normal facial appearance, the proband has typical features of PWS.
Similar content being viewed by others
Introduction
The Prader–Willi syndrome (PWS; MIM 176270) is a rare neurogenetic disorder characterised by decreased fetal and neonatal activity, mild intrauterine growth retardation, severe neonatal hypotonia, hypogenitalism, small hands and feet, facial dysmorphism, hyperphagia and obesity starting at about 18 months, and behavioural disturbances. PWS is caused by a range of genetic defects that disrupt the correct expression of imprinted genes at 15q11–q13. The vast majority of PWS patients (98–99%) present either with a de novo 15q11–q13 deletion affecting the paternally inherited chromosome 15 or with a maternal uniparental disomy for chromosome 15 [upd(15)mat]. Approximately 1% of patients have apparently normal chromosomes 15 of biparental origin, but both chromosomes carry a maternal imprint. These patients have an imprinting defect.
Somatic mosaicism in PWS appears to be very rare. We know of only one case of mosaic upd(15)mat1 and very few cases of mosaic deletions, for example, Chaddha et al2 or mosaic imprinting defects. Among 44 patients with a sporadic imprinting defect, two showed a faint paternal methylation-specific PCR band.3 These patients have not been studied further. The proband described in this study is unique in that she has a high proportion of normal cells.
Proband and methods
Proband
The proposita is the second child of healthy nonconsanguineous parents. At the time of her birth, the mother was 24 years old and the father 29 years. The pregnancy was uneventful, except that fetal movements were reduced in comparison to the older brother; spontaneous birth took place at term and her birth weight was 3500 g. Postnatally, muscular hypotonia and feeding difficulties were noticed, which however did not necessitate tube feeding. Motor milestones were slightly delayed compared to the older brother (unassisted walking with 18 months). The girl was brought up in a bilingual environment during the first 3 years and achieved receptive and expressive skills in both languages. The parents described her as a calm and happy child. Hyperphagia started around the age of 6 years, leading to marked truncal obesity. At about the same time, she started to show temper tantrums. No ritualistic or obsessive-compulsive behaviour was observed, but marked stubbornness was observed. During late childhood, epileptic seizures (absence seizures) were suspected and medication with valproic acid was started but discontinued due to side effects. Except operation for strabismus no major medical interventions were necessary. Learning disabilities necessitated special schooling, and since the age of 18 years she is working and living in a sheltered environment. No information was available about the age when menarche occurred but at present she is suffering from oligomenorrhoea.
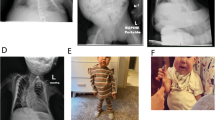
The proband was diagnosed with PWS at the age of 20 years. A detailed clinical evaluation was performed at the age of 21 years (Figure 1): height was 152 cm (<3rd centile) and weight 95 kg (>97th centile), resulting in a body mass index of 41. Head circumference was 52 cm (3rd–10th centile). She showed normal bifrontal diameter, almond-shaped eyes, a thin upper lip, downturned corners of the mouth and a small mandible. In general, the facial appearance was reminiscent of PWS but not typical. No hypopigmentation versus the familial background was observed. Obesity was truncal with abdominal striae. Hand and feet were very small (16 and 20 cm, both below the 3rd centile) with tapering fingers. Multiple lesions from scratching were present on the arms, the legs and on the face.
Microsatellite analysis
DNA was extracted from blood of the proband, her parents, her paternal grandparents and two paternal uncles with standard procedures. A range of microsatellite markers (Research Genetics, Huntsville, AL, USA) was investigated using standard procedures, which map to the critical region on chromosome 15q11–q13. In addition, eight further highly polymorphic markers mapping to different chromosomes were analysed in order to exclude chimerism in the proband.
MSP/DHPLC analysis
MSP/DHPLC analysis was carried out following a previously described method.4, 5 Briefly, DNA modification was carried out using the CpGenome DNA modification Kit (Oncor, Gaithersburg, MD, USA) according to the manufacturer's recommendations using approximately 1 μg genomic DNA. A 250 bp segment of modified DNA was amplified by nested PCR, encompassing a genomic region with differentially methylated CpG positions.4, 5
The DHPLC analysis was performed by loading 8 μl of the PCR products, after heteroduplex formation, on the HPLC (WAVE, Hitachi Model D-7000, Chromatography Data Station Software, Transgenomic LDT Cheshire, UK), using the column DNAsep Cartridge. The running temperature was 59°C and the elution time was 14 min. The starting acetonitrile concentration was 43% with a linear increase to 68% during the first 10 min; 100% acetonitrile was then used for 30 s, and reduced to 43% for the last 4 min.
Restriction analysis
Aliquots of the same 250 bp PCR product that was analysed by MSP/DHPLC was digested with either CfoI or RsaI and visualized on 3% agarose gels. The bands were quantified with the ImageMaster VDS software.
Cytogenetic and fluorescence in situ hybridisation (FISH) investigations
Metaphase chromosome preparations were obtained from PHA-stimulated lymphocyte cultures from the patient according to standard procedures. Conventional GTG-banding was performed at a 400–600 band level according to standard protocols.
Fluorescence in situ hybridisation investigations were performed in metaphase chromosome preparations from the patient using the locus-specific probe LSI Prader–Willi/Angelman region (SNRPN) and control probes CEP15(D15Z1)/PML (15q22) (Vysis, Inc., Downers Grove IL, USA®) according to the manufacturer's recommendations.
Methylation-specific real-time PCR assay
Real-time methylation analysis was performed with a novel PCR assay (Zeschnigk et al,6 Nazlican et al7). In brief, PCR primers were designed to amplify the bisulphite-converted SNURF-SNRPN promoter sequence. An FAM and a VIC-labelled MGB TaqMan® probe specific for the methylated and the unmethylated allele, respectively, were included in each reaction. After calculating the difference of the two CT values within a sample (ΔCT=CT−VIC−CT−FAM), the percentage of methylation was determined with the help of a standard curve. All samples were measured in duplicate, and the mean values are given.
Results
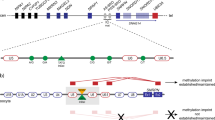
As described previously, differential DNA methylation at the SNURF-SNRPN locus can easily be detected by bisulphite treatment of genomic DNA, followed by PCR and DHPLC analysis. Representative examples of DHPLC curves of individuals with different methylation patterns and the results obtained in our proband are shown in Figure 2.
DHPLC elution curves obtained for control individuals and our proband. The elution curves obtained from PCR products representing a differentially methylated segment of the SNRPN gene are shown for: (a) an unaffected individual; (b) a patient with Angelman syndrome, due to a maternal 15q11–q13 deletion; (c) a PWS patient, due to a paternal 15q11–q13 deletion and (d) the proband described in this work. The peaks representing the nonmethylated (nm) allele elute earlier (between 2.5–3 and 5.5–6.5 min) than the peaks representing the methylated (m) allele (elution time from 6.5 to 7.5 min).
The DHPLC curve of an unaffected individual (Figure 2a) shows two groups of peaks: the PCR product derived from the nonmethylated paternal allele elutes earlier (peaks at 2.5–3 and 5.5–6.5 min) than the PCR product derived from the methylated maternal allele (elution time from 6.5 to 7.5 min). Patients with Angelman or Prader–Willi syndrome typically lack one product. Curve (b) shows the elution profile obtained in a patient with Angelman syndrome due to a maternal 15q11–q13 deletion. Note that only the peaks are observed corresponding to nonmethylated DNA. In contrast, the elution profile obtained in a patient with PWS, due to a paternal 15q11–q13 deletion (curve c), shows only methylated DNA peaks. The pattern obtained in our proband (curve d) is quite different from the other patterns: although peaks representing the two differentially methylated alleles are observed, the nonmethylated allele is clearly less abundant than the methylated allele.
This finding was confirmed by restriction enzyme digestion with CfoI. The restriction fragments were quantified and ratios were calculated between the upper and lower band (representative of the nonmethylated and methylated alleles, respectively). The value in our patient (0.9) is clearly below the values found in normal individuals (1.2–1.5), indicating that our proband has a reduced amount of nonmethylated SNURF-SNRPN PCR alleles. The analysis of other family members (the proband's parents, her two paternal uncles and paternal grandparents) resulted in normal methylation patterns (not shown).
Cytogenetic analysis revealed a normal karyotype (46,XX) in the proband. By FISH analysis of the SNURF-SNRPN locus, we found two signals in each of the 100 metaphases studied. These results excluded a deletion 15q11–q13. Microsatellite analysis revealed maternal and paternal alleles of similar intensity (data not shown). These results confirmed the FISH analysis and excluded upd(15)mat. The proband's paternal alleles at the SNURF-SNRPN locus were found to be derived from the paternal grandmother. There was no indication for chimerism (data not shown). Based on these results, we concluded that the proband is mosaic for an imprinting defect.
The MSP/DHPLC assay allows semiquantitative evaluations owing to the fact that the same pair of primers, in the same PCR reaction, anneal equally efficiently to both methylated and nonmethylated alleles4, 5 (giving rise to same-sized products that differ at about 20 positions within the amplified DNA segment). The ratio of the two products is maintained throughout the PCR, even after a large number of cycles.5 We have previously demonstrated the good semiquantitative aspect of the MSP/DHPLC method based on a patient with a nonmosaic additional invdup15, confirmed by cytogenetic investigations, and a patient with upd(15)mat and a mosaic paternal invdup15, confirmed by FISH analysis.5 This latter patient was also used as a reference in the methylation-specific real-time PCR results presented here (Table 1). The results obtained by FISH analysis, MSP/DHPLC and methylation-specific real-time PCR were in agreement: the presence of the invdup15 chromosome was estimated to occur in 8, 7 and 11% of cells, respectively.
The level of mosaicism in the proband presented in this study was estimated by MSP/DHPLC and methylation-specific real-time PCR. In the former case, we compared the proband's DHPLC elution profile with those obtained by mixing PCR products obtained from modified genomic DNA of normal individuals and PWS patients (data not shown). We estimated that the percentage of normal cells lies between 50 and 60%. The quantitative analysis of the patient's sample by methylation-specific real-time PCR showed very similar results (Table 1): the percentage of methylation was found to be 73%, thus the patient has an equal number of normally methylated cells and abnormal cells.
Discussion
Somatic mosaicism in PWS appears to be very rare. In the previously reported patients with an imprinting defect, the proportion of cells with normally methylated alleles was very low. In contrast, our PWS proband has a relatively high level of normally methylated cells. Nevertheless, she has rather typical PWS. This may indicate that the threshold level of abnormally methylated cells necessary for causing PWS may be less high than assumed. Clearly, in order to determine the clinically relevant threshold level of abnormally methylated cells, it would be necessary to investigate tissues directly involved in the disorder, and to analyse patients with less typical PWS by quantitative assays such as those utilised in this study. The lack of detection of cases with high levels of normally methylated cells may be due in part to milder features, and thus the lack of a clinical diagnosis, and in part to the widespread use of nonquantitative assays for the ascertainment of the methylation status.
There have been several studies on phenotypic differences between patients with PWS due to deletions and those with UPD.8, 9, 10, 11, 12 However, little is known about the specific phenotypic features of individuals with PWS caused by aberrant imprinting.13 Interestingly, distinct features have been reported in some Angelman syndrome patients with imprinting defects.13, 14 The phenotype might also vary according to the degree of mosaicism in different tissues: patients with a higher proportion of normal cells are likely to have a milder phenotype and may escape clinical detection. The largest study of imprinting defects in PWS (reporting on 44 patients) does not include any data on the phenotypes.3 Our patient, although showing aberrant imprinting in only about 50% of peripheral blood cells, showed many classical features of PWS: neonatal hypotonia, childhood-onset obesity, short stature, mild mental retardation and typical behavioural abnormalities (temper tantrums, skin picking, stubbornness). However, she presented with several features which are atypical for PWS: birth weight was normal, there was no need for special feeding in infancy, motor milestones were only slightly delayed and excessive weight gain occurred relatively late (around the age of 6 years). Interestingly, it has been shown that female UPD patients have a shorter course of gavage feeding and a later onset of hyperphagia in comparison to deletion patients, which is reminiscent of the situation in our patient.10 In addition, the facial appearance in our patient is not very typical for PWS (especially the well-defined midface is unusual for classical PWS). Again, it has been reported that the typical facial appearance is more evident in PWS patients with a deletion than in those with UPD.8
In general, it seems that our patient, although presenting with atypical PWS features at birth and in infancy, had progressively acquired more pronounced PWS features during childhood and adolescence. At the time of evaluation (21 years old), she presented with the major signs of PWS, except the relatively normal facial appearance.
The critical paternal 15q11–q13 region in our proband was found to be inherited from the paternal grandmother. This is in agreement with the findings available to date in all the other PWS patients with an imprinting defect and is thought to be indicative of an error in imprint erasure during early spermatogenesis.3 If the imprinting defect in our proband also resulted from such an error, it would be difficult to explain why it is present in a mosaic form. One possibility is that the methylation imprint was only partially erased so that some CpG dinucleotides retained methylation whereas others did not. After fertilisation, this incomplete methylation pattern may have been lost in some cells, whereas in other cells the gaps may have been filled in. However, it is more likely that the imprinting defect occurred after fertilisation, when the genome undergoes massive epigenetic reprogramming. The paternal genome is actively demethylated within the first few hours after fertilisation, and the maternal genome is passively demethylated during subsequent cell divisions. The wave of global demethylation is followed by a wave of global remethylation, which is completed after the blastocyst stage.15 Gametic imprints survive the waves of global de- and remethylation, although it is unclear how they are protected against the global methylation changes. It is possible that the protection against remethylation occasionally fails so that in one cell the paternal SNURF-SNRPN allele is methylated.
References
Horsthemke B, Nazlican H, Husing J et al: Somatic mosaicism for maternal uniparental disomy 15 in a girl with Prader–Willi syndrome: confirmation by cell cloning and identification of candidate downstream genes. Hum Mol Genet 2003; 12: 2723–2732.
Chaddha V, Agarwal S, Phadke SR, Halder A : Low level of mosaicism in atypical Prader–Willi syndrome: detection using fluorescent in situ hybridisation. Indian Pediatr 2003; 40: 166–168.
Buiting K, Gross S, Lich C, Gillessen-Kaesbach G, El-Maarri O, Horsthemke B : Epimutations in Prader–Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am J Hum Genet 2003; 72: 571–577.
Baumer A : Analysis of the methylation status of imprinted genes based on methylation-specific polymerase chain reaction combined with denaturing high-performance liquid chromatography. Methods 2002; 27: 139–143.
Baumer A, Wiedemann U, Hergersberg M, Schinzel A : A novel MSP/DHPLC method for the investigation of the methylation status of imprinted genes enables the molecular detection of low cell mosaicisms. Hum Mutat 2001; 17: 423–430.
Zeschnigk M, Boehringer S, Price EA, Onadim Z, Masshöfer L, Lohmann DR : A novel real-time PCR assay for quantitative analysis of methylated alleles (QAMA): analysis of the retinoblastoma locus. Nucl Acids Res 2004; 32: E125.
Nazlican H, Zeschnigk M, Claussen U et al: Somatic mosaicism in patients with Angelman syndrome and an imprinting defect. Hum Mol Genet 2004; 13: 2547–2555.
Cassidy SB, Forsythe M, Heeger S et al: Comparison of phenotype between patients with Prader–Willi syndrome due to deletion 15q and uniparental disomy 15. Am J Med Genet 1997; 68: 433–440.
Gillessen-Kaesbach G, Robinson W, Lohmann D, Kaya-Westerloh S, Passarge E, Horsthemke B : Genotype–phenotype correlation in a series of 167 deletion and non-deletion patients with Prader–Willi syndrome. Hum Genet 1995; 96: 638–643.
Mitchell J, Schinzel A, Langlois S et al: Comparison of phenotype in uniparental disomy and deletion Prader–Willi syndrome: sex specific differences. Am J Med Genet 1996; 65: 133–136.
Robinson WP, Bottani A, Xie YG et al: Molecular, cytogenetic, and clinical investigations of Prader–Willi syndrome patients. Am J Hum Genet 1991; 49: 1219–1234.
Webb T, Whittington J, Clarke D, Boer H, Butler J, Holland A : A study of the influence of different genotypes on the physical and behavioral phenotypes of children and adults ascertained clinically as having PWS. Clin Genet 2002; 62: 273–281.
Saitoh S, Buiting K, Cassidy SB et al: Clinical spectrum and molecular diagnosis of Angelman and Prader–Willi syndrome patients with an imprinting mutation. Am J Med Genet 1997; 68: 195–206.
Gillessen-Kaesbach G, Demuth S, Thiele H, Theile U, Lich C, Horsthemke B : A previously unrecognised phenotype characterised by obesity, muscular hypotonia, and ability to speak in patients with Angelman syndrome caused by an imprinting defect. Eur J Hum Genet 1999; 7: 638–644.
Reik W, Dean W, Walter J : Epigenetic reprogramming in mammalian development. Science 2001; 293: 1089–1093.
Acknowledgements
We are very grateful to our patient and her family for their willingness to collaborate and for consenting to the publication of the data.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wey, E., Bartholdi, D., Riegel, M. et al. Mosaic imprinting defect in a patient with an almost typical expression of the Prader–Willi syndrome. Eur J Hum Genet 13, 273–277 (2005). https://doi.org/10.1038/sj.ejhg.5201337
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201337
Keywords
This article is cited by
-
Prenatal diagnosis of mosaic chromosomal aneuploidy and uniparental disomy and clinical outcomes evaluation of four fetuses
Molecular Cytogenetics (2023)
-
Prader-Willi syndrome patient with atypical phenotypes caused by mosaic deletion in the paternal 15q11-q13 region: a case report
Italian Journal of Pediatrics (2022)
-
Update of the EMQN/ACGS best practice guidelines for molecular analysis of Prader-Willi and Angelman syndromes
European Journal of Human Genetics (2019)
-
Patients with mosaic methylation patterns of the Prader-Willi/Angelman Syndrome critical region exhibit AS-like phenotypes with some PWS features
Molecular Cytogenetics (2016)