Abstract
Serotonin has been implicated in common disorders involving the central nervous, gastrointestinal, cardiovascular, and pulmonary systems. We describe the first genome-wide screen to identify quantitative trait loci (QTLs) influencing whole blood serotonin in 567 members of a single large pedigree, using a novel association-based mapping approach. We identified an association between the β3 integrin (ITGB3) Leu33Pro polymorphism on 17q21 and whole blood serotonin levels (P-value=9.8 × 10−5). This variant explained the evidence for linkage in this region when included as a covariate in the linkage analysis (change in LOD from 1.87 to 0.16), indicating that ITGB3 may be an important serotonin QTL.
Similar content being viewed by others
Introduction
Serotonin (5-hydroxytryptamine, 5-HT) is typically considered for its role as a brain neurotransmitter; however, it also serves diverse functions in the periphery as a neuromodulator, neurohormone, and vasoactive amine. Peripheral serotonin is produced in the enterochromaffin cells of the intestine and then taken up and stored in platelets, which later release serotonin to modulate vascular constriction and platelet aggregation.1 Disruptions in the peripheral serotonin system have been implicated in a wide range of clinical phenotypes, from cardiac or vascular disorders such as arrhythmias,2 coronary artery disease,3 and migraine headache4 to pulmonary diseases such as asthma5 and pulmonary hypertension.6 Further, whole blood serotonin has been studied as a peripheral marker of brain disorders including autism,7 bipolar disorder,8 Alzheimer's disease,9 and mental retardation10 and of behavioral traits such as aggressi-on11and IQ.12
In a previous study on the heritability of 10 quantitative traits in the Hutterites, a founder population, whole blood serotonin levels had the highest heritability.13 The narrow heritability, which includes only the additive genetic variance, was 0.52, similar to other quantitative traits such as body mass index (BMI) and high-density lipoprotein-cholesterol (HDL-c) levels. However, the broad heritability, which includes all genetic variance, was 0.99. These studies suggested not only that whole blood serotonin levels are highly heritable in the Hutterites, but also that at least one locus in the genome influences serotonin levels in a nonadditive manner (ie, dominant, recessive, or by interaction with other genetic or environmental factors).
Our strategy for identifying serotonin loci begins with a genome-wide analysis of serotonin levels in the Hutterites using recently developed homozygosity linkage and association mapping methods.14 These methods allow us to utilize the inbreeding present in the Hutterites and detect QTLs with recessive main effects. Alone, each method only identifies broad candidate QTL regions. As we hope to identify specific genes that affect serotonin levels, we choose to implement a joint linkage and association analysis. Using joint analysis, we can assess the degree to which a linkage peak identified in the initial genome scan is the result of a particular QTL under that peak. Identifying association to a locus that simultaneously eliminates the linkage peak is a likely indicator of a true QTL.15, 16, 17, 18
We initiated this study to identify the loci that influence whole blood serotonin levels in this founder population. Identifying such loci may not only increase our understanding of the intestinal and platelet serotonin systems, but may also suggest novel susceptibility loci or genetic modifiers for cardiovascular, pulmonary or brain disorders.
Materials and methods
Subjects
The Hutterites of South Dakota, the subjects of our studies, are descendants of only 64 ancestors who lived in the early 1700s to the early 1800s in Europe.19 The usefulness of this population for genetic studies has been described previously.20, 21 A 1623-person pedigree was constructed to include all known ancestors of the 806 individuals in our studies.22 The mean age of the participants was 28.7 years (SD 17.0 years; range 6–89 years). The mean inbreeding coefficient of the individuals in this sample is 0.034 (SD 0.015), slightly greater than that of 1½ cousins.
Our sampling strategy was population-based,20, 23 and enthusiasm for our studies was high, with >95% of colony members participating. As a result, there are no known ascertainment biases that could influence our results. This study was approved by The University of Chicago and University of S Dakota Institutional Review Boards.
Measurement of whole blood serotonin
The blood for measurement of whole blood 5-HT was drawn from 567 Hutterite subjects over 5 years old, after getting informed consent. The samples were immediately transported to the laboratory and stored frozen at −70°C until the time of batch assay. Previous work suggests that diet and/or fasting are unlikely to influence whole blood serotonin levels,24 particularly when diet is as uniform as it is in the Hutterites. While time of sampling may affect whole blood serotonin levels,25 nearly all of the blood sampling was done between 0900 h and noon. Similarly, an influence of seasonal variation on whole blood serotonin levels has been described,25 but samples for these studies were collected during two consecutive winters. Further, the high heritability of whole blood serotonin in this population13 suggests that diet, diurnal variation, and seasonal variation are unlikely to be confounding factors in this study. Assays in platelet-poor plasma revealed that more than 99% of whole blood serotonin is contained in the platelet,26 and whole blood serotonin correlates highly with measures of serotonin per unit number of platelets and serotonin per unit mass of platelet protein.27
Whole-blood 5-HT was analyzed by high-pressure liquid chromatography (HPLC) with fluorometric detection.28 Intra-assay and inter-assay coefficients of variance were 0.8 and 3.6%, respectively. Assays were performed blind to genotypes, family relationships, and all demographic and medical information. The mean whole blood serotonin level in this sample was 191 ng/ml (SD 79). Data were ln-transformed to establish a normal distribution.
Genotyping
A genome screen using 658 autosomal microsatellite markers (Marshfield screening sets 9 and 51) was completed by the Mammalian Genotyping Service of the National Heart, Lung, and Blood Institute, yielding a ∼5-cM map (http://research.marshfieldclinic.org/genetics/). In addition, 226 microsatellite markers and 239 intragenic SNPs or in/dels related to asthma and cardiovascular diseases (eg see Newman et al29) were genotyped in this sample. None of the markers were selected because they were functional or positional ‘candidates’ for serotonin levels. Genotyping was performed blind to all phenotypic information. Distances for framework markers are based on the Marshfield map (http://research.marshfieldclinic.org/genetics/); all other markers were placed using the physical map (http://genome.ucsc.edu/) and estimations of recombination within the Hutterite pedigree by CRI-MAP (http://biobase.dk/Embnetut/Crimap/). The final map had an average intermarker distance of 3.2 cM.
Statistical analysis
Genome scans were performed using HBD linkage and association methods, and significance was assessed using a permutation-based method. The linkage method tests for correlations between regions inherited homozygous by descent (HBD) and trait value, while the association method, allele-specific HBD (ASHBD) tests for correlations between specific alleles at markers inherited HBD and trait value. These methods are the only multipoint methods available capable of analyzing the Hutterite pedigree unbroken and are described in detail elsewhere.14 For both methods, we assess empirical locus-specific and genome-wide significance using a Monte Carlo permutation test. This test keeps the genotypes fixed while permuting trait values. This has the advantage of assessing significance conditional on characteristics of the genotype data (eg informativeness, heterozygosity, linkage disequilibrium, etc), while preserving the covariance structure in the phenotype data. Details of how this is accomplished can be found in Abney et al.14 In addition to the empirical P-values, the locus-specific P-values were Bonferroni corrected to adjust for multiple tests when several alleles were present at a marker locus. The Bonferroni-corrected P-values, which were very close to the locus-specific permutation-based P-values, are reported here. In addition, for each of the P-values, we report the equivalent one degree of freedom LOD score. Suggestive significance was met if the P-value at a locus was less than the expected minimum P-value under the null hypothesis.
In the covariate analyses, the probability of HBD for a specific allele at a marker is used as a covariate in the linkage analysis. Thus, we can determine the residual linkage at a locus after removing effects of the association. This approach has been the focus of recently developed methods aimed at identifying the specific genes and polymorphisms that produce an observed linkage peak (eg Cardon and Abecasis,15 Fulker et al,16 Soria et al,17 and Sun et al18).
Results
Association analysis by allele-specific homozygosity-by-descent (ASHBD)
Using the multipoint ASHBD association method14 for whole blood serotonin level, two loci reached the threshold for suggestive significance (see Materials and methods). Results with P-value <0.01 are shown in Table 1. The Leu33 allele at the β3 integrin (ITGB3) locus on 17q21 at 66 cM from P-terminus (p-ter) provided the strongest signal and was associated with lower levels of whole blood serotonin levels (P-value=9.8 × 10−5, genome-wide P-value=0.21). Three other alleles at flanking loci between 57.6 and 66.3 cM on chromosome 17 showed some evidence for association, with P-values <0.01. ITGB3 is about 17 cM distal to the serotonin transporter locus (SLC6A4), which showed only a modest association with serotonin levels in this study (P-value=0.015). A second locus, the vitamin D receptor (VDR) on chromosome 12q12–q14, also met the criteria for suggestive significance (P-value=2.4 × 10−4). However, the association of whole blood serotonin levels with VDR can be explained by a primary association of VDR with platelet count (data not shown). The association with ITGB3 has no primary association with platelet count, and an association with serotonin levels remains after accounting for platelet number (data not shown).
Linkage analysis by locus-specific HBD
Using the HBD linkage method,14 one locus reached the threshold for suggestive linkage (Figure 1). This signal was on chromosome 16p13-p12 at 35 cM from p-ter (LOD=2.5, P-value=0.0007). The peak included the locus D16S764, at which homozygosity for the 108 bp allele was also associated with higher serotonin levels by the ASHBD test (P-value=0.008). The second largest linkage peak was on chromosome 17 at 70 cM, near the ITGB3 locus (LOD=1.87, P-value=0.0033).
Covariate analysis by locus-specific HBD
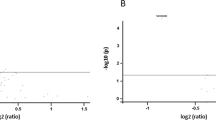
To determine whether HBD for the Leu33 allele at the ITGB3 locus accounted for the linkage signal on chromosome 17q, we repeated the HBD linkage analysis including probability of HBD for the Leu33 allele as a covariate. The variation in whole blood serotonin levels attributable to HBD for the Leu33 allele at the ITGB3 locus is removed, and evidence for linkage with the remaining variation is evaluated. In the covariate analysis, the linkage peak in the ITGB3 region on 17q was nearly completely eliminated (LOD=0.16, P-value=0.39 at 67 cM; Figure 2). Thus, HBD for the Leu33 allele, or other variants that are in linkage disequilibrium (LD) with it, explained all of the evidence for linkage on chromosome 17q.
Covariate linkage analysis of whole blood serotonin for chromosome 17. The solid line is the original linkage signal (Figure 1) and the dashed line indicates the linkage results with ITGB3 Leu33Pro as a covariate. Locus-specific P-values are shown on the left axis and LOD equivalents on the right axis.
Discussion
This is the first genome-wide association or linkage study of the quantitative trait whole blood serotonin. Our study was conducted in a sample that was unselected with respect to any particular phenotype, allowing us to study normal levels of variation in this important signaling molecule. Further, the reduced environmental variation resulting from this population's communal lifestyle should enhance the relative influence of genetics on variation in a quantitative trait. This is likely reflected in the high heritability of whole blood serotonin in the Hutterite population, although this does not necessarily correspond to simple inheritance or even the presence of a major QTL.20 However, the association with the Leu33Pro polymorphism in ITGB3, when inherited HBD, was strong (P=9.8 × 10−5) and accounted for nearly all the evidence for linkage in the region, indicating that this polymorphism lies on a haplotype that acts as a recessive QTL for whole blood serotonin. In addition to the ITGB3 locus, this study identified a linkage signal on chromosome 16p that coincides with linkage peaks for autism30 and attention-deficit hyperactivity disorder.31
ITGB3 encodes a β-integrin that comprises part of the platelet-specific heterodimeric receptors for fibrinogen and the widely expressed heterodimeric receptors for vitronectin. Fibrinogen receptors regulate platelet aggregation, hemostasis, and thrombosis. In tissues other than platelets, integrin activation of kinases and small GTP-binding proteins through vitronectin receptors is critical for processes such as cell migration, proliferation, survival, differentiation, and synapse maturation.32, 33, 34
The ITGB3 Leu33Pro polymorphism determines the platelet-specific antigen PlA1/PlA2 (corresponding to Leu33/Pro33, respectively). The protein carrying the PlA2 allele has been shown to have increased and more stable interactions with fibrinogen,35 enhanced platelet aggregability and thrombin generation,36, 37, 38 and enhanced cell migration.39 Further, the PlA2 allele has been associated with coronary thrombosis,40, 41 stroke,42, 43 and myocardial infarction.44, 45 Studies of ITGB3 polymorphisms and whole blood serotonin in patients with cardiovascular diseases and controls will be required to evaluate whether altered serotonin levels mediate the association between PlA2 and cardiovascular phenotypes. In fact, because previous association studies of ITGB3 considered only the Leu33Pro polymorphism, it is possible that the associated Leu33 allele in some of these earlier studies, as well as in this study, is a marker for other variation in the gene.
In summary, we identified a novel association between a polymorphism in ITGB3 and whole blood serotonin levels, using methods that harness the extensive linkage disequilibrium in this founder population. A direct connection between platelet integrin function and whole blood serotonin has not previously been noted, but our results suggest that integrin signaling may affect the uptake, storage, or release of serotonin in the platelet. Lastly, this study identifies the ITGB3 gene as a tool for further genetic dissection of the serotonin signaling pathway and as a candidate for other common disorders that have been associated with the serotonin system.
References
Frishman WH, Grewall P : Serotonin and the heart. Ann Med 2000; 32: 195–209.
Pino R, Cerbai E, Calamai G et al: Effect of 5-HT4 receptor stimulation on the pacemaker current I(f) in human isolated atrial myocytes. Cardiovasc Res 1998; 40: 516–522.
Nair GV, Gurbel PA, O'Connor CM, Gattis WA, Murugesan SR, Serebruany VL : Depression, coronary events, platelet inhibition, and serotonin reuptake inhibitors. Am J Cardiol 1999; 84: 321–323, A328.
Peroutka SJ, McCarthy BG : Sumatriptan (GR 43175) interacts selectively with 5-HT1B and 5-HT1D binding sites. Eur J Pharmacol 1989; 163: 133–136.
Cazzola I, Matera MG : 5-HT modifiers as a potential treatment of asthma. Trends Pharmacol Sci 2000; 21: 13–16.
Egermayer P, Town GI, Peacock AJ : Role of serotonin in the pathogenesis of acute and chronic pulmonary hypertension. Thorax 1999; 54: 161–168.
Cook E, Leventhal B : The serotonin system in autism. Curr Opin Pediatr 1996; 8: 348–354.
Wirz-Justice A, Puhringer W : Increased platelet serotonin in bipolar depression and hypomania. J Neural Transm 1978; 42: 55–62.
Kumar AM, Sevush S, Kumar M, Ruiz J, Eisdorfer C : Peripheral serotonin in Alzheimer's disease. Neuropsychobiology 1995; 32: 9–12.
Partington MW, Tu JB, Wong CY : Blood serotonin levels in severe mental retardation. Develop Med Child Neurol 1973; 15: 616–627.
Moffitt T, Brammer G, Caspi A et al: Whole blood serotonin relates to violence in an epidemiological study. Biol Psychiatry 1998; 43: 446–457.
Cook EH, Leventhal BL, Heller W, Metz J, Wainwright M, Freedman DX : Autistic children and their first-degree relatives: relationships between serotonin and norepinephrine levels and intelligence. J Neuropsychiatr Clin Neurosci 1990; 2: 268–274.
Abney M, McPeek MS, Ober C : Broad and narrow heritabilities of quantitative traits in a founder population. Am J Hum Genet 2001; 68: 1302–1307.
Abney M, Ober C, McPeek MS : Quantitative trait homozygosity and association mapping and empirical genome-wide significance in large complex pedigrees: fasting serum insulin levels in the Hutterites. Am J Hum Genet 2002; 70: 920–934.
Cardon LR, Abecasis GR : Some properties of a variance components model for fine-mapping quantitative trait loci. Behav Genet 2000; 30: 235–243.
Fulker DW, Cherny SS, Sham PC, Hewitt JK : Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet 1999; 64: 259–267.
Soria JM, Almasy L, Souto JC et al: Linkage analysis demonstrates that the prothrombin G20210A mutation jointly influences plasma prothrombin levels and risk of thrombosis. Blood 2000; 95: 2780–2785.
Sun L, Cox NJ, McPeek MS : A statistical method for identification of polymorphisms that explain a linkage result. Am J Hum Genet 2002; 70: 399–411.
Ober C, Weitkamp LR, Cox N, Dytch H, Kostyu D, Elias S : HLA and mate choice in humans. Am J Hum Genet 1997; 61: 497–504.
Ober C, Abney M, McPeek MS : The genetic dissection of complex traits in a founder population. Am J Hum Genet 2001; 69: 1068–1079.
Abney M, Ober C, McPeek MS : Homozygosity mapping of quantitative trait loci in complex inbred pedigrees. Am J Hum Genet 2000; 67: 327.
Abney M, McPeek M, Ober C : Estimation of variance components of quantitative traits in inbred populations. Am J Hum Genet 2000; 66: 629–650.
Ober C, Tsalenko A, Parry R, Cox NJ : A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet 2000; 67: 1154–1162.
Cook EH, Anderson G, Heninger G, Fletcher KE, Freedman DX, Leventhal BL : Tryptophan loading in hyperserotonemic and normoserotonemic adults. Biol Psychiatry 1992; 31: 525–528.
Wirz-Justice A, Lichtsteiner M, Feer H : Diurnal and seasonal variations in human platelet serotonin in man. J Neural Transm 1977; 41: 7–15.
Anderson GM, Feibel FC, Cohen DJ : Determination of serotonin in whole blood, platelet-rich plasma, platelet-poor plasma and plasma ultrafiltrate. Life Sci 1987; 40: 1063–1070.
Jernej B, Banovic M, Cicin-Sain L et al: Physiological characteristics of platelet/circulatory serotonin: study on a large human population. Psychiatry Res 2000; 94: 153–162.
Anderson GM, Young JG, Cohen DJ, Schlicht KR : Liquid-chromatographic determination of serotonin and tryptophan in whole blood and plasma. Clin Chem 1981; 27: 775–776.
Newman D, Hoffjan S, Bourgain C et al: Are common disease susceptibility alleles the same in outbred and founder populations?, 2004, in press.
International Molecular Genetic Study of Autism Consortium: A genome wide screen for autism: strong evidence for linkage to chromosomes 2q, 7q and 16p. Am J Hum Genet 2001; 69: 570–581.
Smalley SL, Kustanovich V, Minassian SL et al: Genetic linkage of attention-deficit/hyperactivity disorder on chromosome 16p13, in a region implicated in autism. Am J Hum Genet 2002; 71: 959–963.
Bodeau AL, Berrier AL, Mastrangelo AM, Martinez R, LaFlamme SE : A functional comparison of mutations in integrin beta cytoplasmic domains: effects on the regulation of tyrosine phosphorylation, cell spreading, cell attachment and beta1 integrin conformation. J Cell Sci 2001; 114: 2795–2807.
Chavis P, Westbrook G : Integrins mediate functional pre- and postsynaptic maturation at a hippocampal synapse. Nature 2001; 411: 317–321.
Kramar EA, Bernard JA, Gall CM, Lynch G : Integrins modulate fast excitatory transmission at hippocampal synapses. J Biol Chem 2003; 10: 10.
Goodall AH, Curzen N, Panesar M et al: Increased binding of fibrinogen to glycoprotein IIIa-proline33 (HPA-1b, PlA2, Zwb) positive platelets in patients with cardiovascular disease. Eur Heart J 1999; 20: 742–747.
Feng D, Lindpaintner K, Larson MG et al: Increased platelet aggregability associated with platelet GPIIIa PlA2 polymorphism: the Framingham Offspring Study. Arterioscler Thromb Vasc Biol 1999; 19: 1142–1147.
Vijayan KV, Goldschmidt-Clermont PJ, Roos C, Bray PF : The Pl(A2) polymorphism of integrin beta(3) enhances outside-in signaling and adhesive functions. J Clin Invest 2000; 105: 793–802.
Undas A, Brummel K, Musial J, Mann KG, Szczeklik A : Pl(A2) polymorphism of beta(3) integrins is associated with enhanced thrombin generation and impaired antithrombotic action of aspirin at the site of microvascular injury. Circulation 2001; 104: 2666–2672.
Sajid M, Vijayan KV, Souza S, Bray PF : PlA polymorphism of integrin beta 3 differentially modulates cellular migration on extracellular matrix proteins. Arterioscler Thromb Vasc Biol 2002; 22: 1984–1989.
Weiss EJ, Bray PF, Tayback M et al: A polymorphism of a platelet glycoprotein receptor as an inherited risk factor for coronary thrombosis. N Engl J Med 1996; 334: 1090–1094.
Mikkelsson J, Perola M, Laippala P, Penttila A, Karhunen PJ : Glycoprotein IIIa Pl(A1/A2) polymorphism and sudden cardiac death. J Am Coll Cardiol 2000; 36: 1317–1323.
Wagner KR, Giles WH, Johnson CJ et al: Platelet glycoprotein receptor IIIa polymorphism P1A2 and ischemic stroke risk: the stroke prevention in young women study. Stroke 1998; 29: 581–585.
Carter AM, Catto AJ, Bamford JM, Grant PJ : Platelet GP IIIa PlA and GP Ib variable number tandem repeat polymorphisms and markers of platelet activation in acute stroke. Arterioscler Thromb Vasc Biol 1998; 18: 1124–1131.
Pastinen T, Perola M, Niini P et al: Array-based multiplex analysis of candidate genes reveals two independent and additive genetic risk factors for myocardial infarction in the Finnish population. Hum Mol Genet 1998; 7: 1453–1462.
Carter AM, Ossei-Gerning N, Wilson IJ, Grant PJ : Association of the platelet Pl(A) polymorphism of glycoprotein IIb/IIIa and the fibrinogen Bbeta 448 polymorphism with myocardial infarction and extent of coronary artery disease. Circulation 1997; 96: 1424–1431.
Acknowledgements
Sabine Hoffjan, Raluca Nicolae, Elle Profits, Dan Schneider, Robert Stanaker, Shuya Yan, and Zhi-Ying Yang provided technical assistance. Michael Grow, Arkadiy Silbergleit, Lori Steiner, and Karen Walker helped develop some of the SNP genotyping reagents. This study was supported in part by grants P50 HL56399 and R01 HL66533 (CO), R01 HG001645 (MSM), and K02 MH01389 (EC), and RDK551 (MA) the NHLBI Mammalian Genotyping Service (CO), and a grant from Hoffmann-LaRoche, Inc (CO). LAW was supported by a NSF graduate research fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Weiss, L., Veenstra-VanderWeele, J., Newman, D. et al. Genome-wide association study identifies ITGB3 as a QTL for whole blood serotonin. Eur J Hum Genet 12, 949–954 (2004). https://doi.org/10.1038/sj.ejhg.5201239
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201239
Keywords
This article is cited by
-
Experimental Models for Autism Spectrum Disorder Follow-Up for the Validity
Review Journal of Autism and Developmental Disorders (2016)
-
Integrin β3 Haploinsufficiency Modulates Serotonin Transport and Antidepressant-Sensitive Behavior in Mice
Neuropsychopharmacology (2015)
-
PLA1A2 platelet polymorphism predicts mortality in prediabetic subjects of the population based KORA S4-Cohort
Cardiovascular Diabetology (2014)
-
Neuroimaging Endophenotypes in Animal Models of Autism Spectrum Disorders: Lost or Found in Translation?
Psychopharmacology (2014)
-
Genetic background modulates phenotypes of serotonin transporter Ala56 knock-in mice
Molecular Autism (2013)