Abstract
The endoplasmic reticulum (ER) has evolved specific mechanisms to ensure protein folding as well as the maintenance of its own homeostasis. When these functions are not achieved, specific ER stress signals are triggered to activate either adaptive or apoptotic responses. Here, we demonstrate that MCF-7 cells are resistant to tunicamycin-induced apoptosis. We show that the expression level of the ER chaperone calnexin can directly influence tunicamycin sensitivity in this cell line. Interestingly, the expression of a calnexin lacking the chaperone domain (ΔE) partially restores their sensitivity to tunicamycin-induced apoptosis. Indeed, we show that ΔE acts as a scaffold molecule to allow the cleavage of Bap31 and thus generate the proapoptotic p20 fragment. Utilizing the ability of MCF-7 cells to resist tunicamycin-induced apoptosis, we have characterized a molecular mechanism by which calnexin regulates ER-stress-mediated apoptosis in a manner independent of its chaperone functions but dependent of its binding to Bap31.
Similar content being viewed by others
Main
The endoplasmic reticulum (ER) is an organelle specialized for folding and assembly of secretory proteins. In such a calcium-rich and oxidizing environment, nascent polypeptide chains can be modified by addition of N-linked oligosaccharides and formation of intra- and inter-disulfide bonds. Several ER resident proteins such as, BiP/Grp78, Grp94, PDI, calreticulin and calnexin act as molecular chaperones to promote proper folding and/or prevent aggregation of nascent protein chains.1 In addition, this compartment also contains the molecular machinery necessary to ensure the quality control and degradation of terminally misfolded proteins as well as signalling components that mediate communications between the ER and the rest of the cell.2 The ER is also involved in the regulation of the intrinsic apoptotic pathway through various mechanisms.3 These proapoptotic pathways are induced by chronic stresses applied to the ER.4
Treatment of cells with the antibiotic tunicamycin, which prevents the transfer of the oligosaccharide chain from the dolichol-phosphate donor to specific asparagine residues on the nascent polypeptide chain,5 leads to the accumulation of misfolded nonglycosylated proteins in the ER. This saturation of the ER folding capacity, induces in turn the activation of the ER-specific adaptive response: the Unfolded Protein Response.4 When the inhibition of N-glycosylation is maintained, cells are then unable to restore the homeostasis of the ER and undergo an apoptotic process. It was previously reported in the literature that several cancer cell lines were resistant to tunicamycin-induced apoptosis. More specifically, the human breast carcinoma cell line MCF-7 has been shown to tolerate long-term tunicamycin treatments.6 This was suggested to result from the deregulation of the molecular chaperone expression,6 however, no demonstration of this hypothesis has been obtained so far.
Inhibition of N-glycosylation by tunicamycin prevents the association of nascent N-glycoprotein chains with calnexin.5 This protein is an ER type I integral membrane chaperone which transiently binds to newly synthesized mono-glucosylated glycoproteins, thereby promoting their folding and oligomerization.1, 7, 8 Recently, calnexin has been reported to be involved in the mediation of late apoptotic processes following prolonged ER stresses through its caspase-mediated cleavage9 or its association with the ER resident caspase-8 substrate, Bap31.10
In this study, we investigate whether calnexin expression levels can directly influence tunicamycin sensitivity in the human breast cancer cell line MCF-7. We demonstrate that calnexin expression level is an important regulator of tunicamycin-induced apoptosis. In addition, we show that a deletion in the chaperone domain of calnexin (ΔE) leads to the restoration of the sensitivity to tunicamycin and correlates with the destabilization of the complex formed by calnexin and Bap31. These results suggest that calnexin may regulate tunicamycin-induced apoptosis pathways in a manner dependent on Bap31 as a scaffold molecule to allow Bap31 cleavage by caspase-8, thus generating its proapoptotic product Bap20.
Results
Calnexin expression in tunicamycin resistant cells
To evaluate the responsiveness of various cell lines to tunicamycin, we treated eight tumor and nontumor cell lines (Rat2, COS7, MDCK, Jurkat, HuH7, Hela, HepG2 and MCF-7) with tunicamycin (10 μg/ml) for 24 h and measured the resulting numbers of apoptotic cells. Using TUNEL assays (Figure 1a), we determined the levels of apoptosis in each cell line and found approximately 15–30% of apoptotic cells after 24 h of tunicamycin treatment in seven out of eight cell lines (Rat2, COS7, MDCK, Jurkat, HuH7, Hela and HepG2) (Figure 1a). Surprisingly, MCF-7 cells showed a reduced sensitivity to tunicamycin-induced apoptosis and exhibited apoptosis levels of approximately 5% (Figure 1a). In an attempt to identify proteins potentially involved in this resistance, we examined the global changes in protein expression in the presence or not of tunicamycin in MCF-7. To this end, cells were incubated (+) or not (−) with tunicamycin for 24 h and cells extracts were resolved by SDS–PAGE and stained with Coomassie blue. A reproducible separation of the protein extracts revealed an intense band migrating at approximately 90 kDa, whose intensity increased by approximately two- to three folds upon tunicamycin treatment (Figure 1b, arrow). This band was then excised from the gel, trypsin-digested and the resulting peptides analyzed by mass spectrometry. MS/MS analysis of these peptides revealed that the most prominent protein in this band was calnexin. A representative MS/MS spectra corresponding to a peptide specific of calnexin is shown in Figure 1c. Additionally, to confirm calnexin upregulation upon tunicamycin treatment in MCF-7 cells, the same extracts were immunoblotted using anticalnexin (CNX) antibodies (Figure 1d) and as expected, calnexin expression levels increased in response to tunicamycin treatment (as compared to the expression of the cytosolic MAPK, ERK-1). We next examined the changes in calnexin expression in the presence or not of tunicamycin in the various cell lines mentioned above (Rat2, COS7, Jurkat, Hela and HepG2). Cells were incubated (+) or not (−) with tunicamycin for 24 h and cells extracts were resolved by SDS–PAGE followed by immunoblotting using CNX antibodies (Figure 1e). Calnexin expression levels did not show any significant variation in all the cell lines tested. These data indicate that the calnexin expression is specifically increased in MCF-7 cell line upon tunicamycin treatment. As tunicamycin-mediated inhibition of N-glycosylation prevents the association of nascent glycoprotein chains with calnexin, MCF-7 cells were pulse-labelled with 35S-Label for 15 min with or without a 90 min pretreatment with tunicamycin and the association of newly synthesized proteins with calnexin was then assessed by immunoprecipitation of calnexin and separation of the immunoprecipitates by SDS–PAGE followed by radioautography. As expected, we found that tunicamycin treatment efficiently abrogated calnexin association with newly synthesized substrates (data not shown), thus suggesting that the observed resistance of MCF-7 to tunicamycin-induced apoptosis may be independent of glycoprotein binding to calnexin luminal domain.
Characterization of tunicamycin effect on MCF-7 cells. (a) Rat2, COS7, MDCK, Jurkat, HuH7, Hela, HepG2 and MCF-7 cells were treated for 24 h with 10 μg/ml tunicamycin in DMEM containing 0.2% FBS. Floating and attached cells were collected, fixed in 100% ice-cold methanol and the number of apoptotic cells determined by a TUNEL assay. (b) MCF-7 were incubated (+) or not (−) with 10 μg/ml tunicamycin for 24 h. Cells extracts were resolved by SDS–PAGE and stained with Coomassie blue. (c) The relevant band in Figure 1b was subjected to digestion by trypsin and treated as described in Materials and methods. Tryptic peptides were analyzed on MS/MS mode and results analyzed using the Mascot Software. A typical peptide mass spectrum is shown as a representative result. (d) MCF-7 were incubated (+) or not (−) with 10 μg/ml tunicamycin for 24 h. Cells extracts were either directly immunoblotted with anti-ERK-1 and anti-calnexin (CNX) antibodies. (e) Rat2, COS7, Jurkat, Hela and HepG2 cells were incubated (+) or not (−) with 10 μg/ml tunicamycin for 24 h. 20 μg of each cells extract were directly immunoblotted with anti-calnexin (CNX) antibodies
Effects of tunicamycin on MCF-7 cells expressing caspase-3
MCF-7 cells are deficient in caspase-3, an effector caspase, which plays an important role in the execution of apoptosis. Thus, caspase-3 deficiency may potentially contribute to the observed tunicamycin resistance of MCF-7. To investigate the specific contribution of caspase-3 in tunicamycin-induced apoptosis, we have compared the responsiveness to tunicamycin of parental MCF-7 cells (Mock; caspase-3 deficient) to that of MCF-7 cells transfected with a caspase-3 expression vector (Casp3) (Figure 2a). Using a viability assay (MTT assay), we show that tunicamycin treatment of MCF-7 Casp3 cells led to biological effects similar to those observed in MCF-7 cells (Mock) (Figure 2b). To verify whether tunicamycin treatment affected cell growth and not the metabolic rate in MCF-7 Casp3 cells, an apoptosis assay (TUNEL) was also carried out. As shown in Figure 2c, tunicamycin treatment led to a similar effect in MCF-7 cells (Mock) and MCF-7 Casp3 cells which both exhibited approximately only 5% of apoptosis. These data show that MCF-7 resistance to tunicamycin is not related MCF-7 caspase deficiency. In NIH3T3 cells, calnexin was found to be cleaved upon treatment with inducers of apoptosis and ER stress (such as tunicamycin). The resulting cleavage products were shown to be involved in apoptosis resistance mechanisms.9 As the cleavage of calnexin resulted in apoptosis attenuation, we tested whether calnexin was cleaved in MCF-7 Casp3 and MCF-7 cells in response to tunicamycin (Figure 2d). Cells were incubated (+) or not (−) with tunicamycin for 24 h and the corresponding cell extracts were immunoblotted using two CNX antibodies raised against two synthetic peptides corresponding to amino acids 30–48 in the luminal domain (α CNX-C1) and 555–573 in the cytoplasmic domain (α CNX-C4) respectively.11 Using these two antibodies, we demonstrated that calnexin was cleaved neither in MCF-7 Casp3 nor in MCF-7 cells under basal- or tunicamycin-treated conditions (Figure 2d). These results indicate that the cytoplasmic domain of calnexin is not cleaved by tunicamycin in our experimental system and thus, is not likely to participate in the observed resistance to tunicamycin-induced apoptosis (Figure 2c).
Characterization of tunicamycin effects on MCF-7 cells expressing caspase-3. (a) MCF-7 cells were transfected by Lipofectamine 2000 with the empty vector CMV (Mock) or the same vector containing the Myc-caspase-3 protein (Casp3). The presence of caspase-3 was assessed by immunoprecipitation of Mock or Casp3 cell lysates using anti-Myc followed by immunoblotting with anti-Myc antibody. (b) MCF-7 cells were grown in Dulbecco's Modified Eagle Medium (DMEM) containing 10% FBS over a 1-day period in the presence or absence of 10 μg/ml tunicamycin. Cell proliferation was assessed by MTT colorimetric assay in triplicates and (c) the number of apoptotic cells was determined by a TUNEL assay. Values are expressed in arbitrary units. (d) MCF-7 cells, transfected with a CMV promoter containing vector (Mock) or the same vector containing the Myc-tagged caspase-3 cDNA (Casp3), were incubated (+) or not (−) with 10 μg/ml tunicamycin for 24 h. Then, cells extracts were directly immunoblotted with different anti-calnexin antibodies (CNX-C1 and CNX-C4) and anti-ERK-1 antibody
Effects of tunicamycin on calnexin protein expression in MCF-7R
To study the potential mechanisms responsible for their acquired resistance to tunicamycin, MCF-7 cells clones were selected for their ability to grow in presence of this antibiotic as illustrated in Figure 3a. We isolated three clones (MCF-7R (1–3)) resistant to tunicamycin in the conditions described in Materials and Methods. As shown in Figure 3b, MCF-7R resistant cell lines (the experiment shown was carried out on MCF-7R1 and repeated on R2 and R3) were not targeted to apoptosis after 24 h treatment with tunicamycin as assessed by TUNEL analysis. As calnexin expression levels increased upon tunicamycin treatment in the MCF-7 parental cells, we tested whether calnexin expression level was differentially altered in response to tunicamycin in MCF-7R, as compared to MCF-7. Using an immunoblotting approach, we demonstrated that the induction of calnexin expression in response to tunicamycin was smaller in MCF-7R compared to MCF-7 cells (Figure 3c). However under basal conditions, calnexin expression levels were approximately two- to three folds higher in MCF-7R than in MCF-7 cells. These observations suggest that calnexin expression levels may regulate tunicamycin sensitivity in MCF-7 cells.
Establishment and characterization of tunicamycin effect on a tunicamycin-resitant MCF-7 cell line. (a) Schematic representation of the procedure used to establish the tunicamycin-resistant cell lines: MCF-7R (1–3). Step 0: Addition of 10 μg/ml tunicamycin to a MCF-7 cell culture; Step 1: Emergence of colonies; Step 4: Distinct colony transferred to a T25 flask; Step 10: Weekly split ratio of 1 : 10; Step 12: Weekly split ratio of 1 : 20. (b) MCF-7 and MCF-7R were passaged at 3 × 103 cells/cm2 in DMEM containing 10% FBS without tunicamycin. Two days later, MCF-7 and MCF-7R cells were treated for 24 h with 10 μg/ml tunicamycin. The number of apoptotic cells, determined by TUNEL assay, was expressed as a percentage of the total input. The graph shows the mean of three independent experiments. (c) MCF-7 and MCF-7R cells were treated in the same experimental conditions as described in b and incubated (+) or not (−) with 10 μg/ml tunicamycin for 24 h. Cells extracts were either directly immunoblotted with anti-ERK-1 and anti-calnexin (CNX) antibodies. Results are the representative of three independent experiments
Inhibition of calnexin expression increases tunicamycin sensitivity in MCF-7 cells
To determine whether a decrease in calnexin expression in MCF-7 cells can modulate their sensitivity to tunicamycin, we analyzed the levels of tunicamycin-induced apoptosis in cells exhibiting a siRNA-mediated reduced expression of calnexin. Cell lysates from MCF-7 cells transfected with siRNA against the human calnexin (either S2 or S3) were analyzed by immunoblot (Figure 4a) and showed the loss of over 70 and 55% of total cellular calnexin protein expression after 48 h in culture, respectively, for S2 and S3. Figure 4a also shows that both siRNA effectively decreased specifically calnexin expression without affecting calnexin closest homolog calreticulin (middle panel, CRT). To evaluate the effect of calnexin silencing on sensitivity to tunicamycin-induced apoptosis, a 24 h tunicamycin treatment was carried out on MCF-7 and MCF-7R cells transfected with either scrambled (Scr) or siRNA calnexin (S2) and the percentage of apoptotic cells was then evaluated (Figure 4b and d, as assessed by TUNEL assay). The previously described tunicamycin-dependent induction of calnexin expression (Figures 1d and 3c) was also observed in MCF-7 cells silenced for calnexin (MCF-7+S2) but to a lesser extend than in MCF-7 cells transfected with a scrambled siRNA (MCF-7+Scr) (Figure 4c). Calnexin expression levels decreased in MCF-7R cell transfected with siRNA calnexin (MCF-7R+S2) compared to MCF-7R cells transfected with scrambled siRNA (MCF-7R+Scr) (Figure 4e). As expected, a lower expression of calnexin correlated with a larger number of apoptotic cells observed upon tunicamycin treatment (Figure 4b and c, S2 as compared to Scr). To assess whether calnexin silencing increased the sensitivity of MCF-7 cells to other apoptotic agents, we compared drug sensitivity of MCF-7 calnexin silenced (MCF-7+S2) to that of the nonsilenced cells (MCF-7+Scr) using a MTT assay. As shown in Figure 4f, the sensitivity of the MCF-7 cells to dithiotreitol (DTT) was increased significantly upon calnexin silencing. In parallel, the same experiment was carried out in MCF-7R cells and indicated that these cells were also resistant to DTT-induced apoptosis. In contrast, the sensitivity to other drugs (Azetidine-2-Carboxylic Acid: AZC; Gentamicin: Gen; and Herbimycin A: Herb) did not significantly vary in the different cell types (MCF-7, MCF-7 S2 and MCF-7R). Interestingly, the effects of tunicamycin and DTT on apoptosis resistance in MCF-7, MCF-7 S2 and MCF-7R cells were similar, thus suggesting a specific effect on the ER folding machinery.
Effect of siRNA calnexin on sensitivity of MCF-7 to tunicamycin. (a) Lysates of MCF-7 cells 3 days after transfection with calnexin siRNA duplexes (S2 and S3) or nontransfected (C) were immunoblotted using anti-calnexin (CNX), anti-ERK-1 and anti-calreticulin (CRT) antibodies. (b) MCF-7R and MCF-7 transfected with calnexin (S2) or scrambled (scr) siRNA duplexes were treated for 24h with 10 μg/ml tunicamycin. The number of apoptotic cells, determined by TUNEL assay, was expressed as a percentage of the total input. The graph shows the mean of three independent experiments. (c) MCF-7 transfected with calnexin (MCF-7 S2) or scrambled (Scr) siRNA duplexes were incubated (+) or not (−) with 10 μg/ml tunicamycin for 24 h. Cells extracts were directly immunoblotted using either anti-ERK-1 or anti-calnexin (CNX) antibodies. Results are the representative of three independent experiments. Same experiments were repeated with the MCF-7R cell line in panels d and e. These experiments were repeated three times. (f) MCF-7, MCF-7 S2 and MCF-7R cells were treated or not (Ctl) for 24h with 10 μg/ml tunicamycin (Tun), 10 mM Azetidine-2-Carboxylic Acid (AZC), 10mM dithiotreitol (DTT), 50mM Genistein (Gen) and 50 mM Herbimycin A (Herb). Then, cell proliferation was assessed by MTT colorimetric assay in triplicates
Characterization of the calnexin construct ΔE
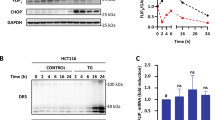
Inhibition of N-glycosylation by tunicamycin prevents the association of nascent N-glycoprotein chains with calnexin.5 Calnexin is an endoplasmic reticulum protein with a lectin-like chaperone function contained in its luminal domain and which transiently binds to newly synthesized mono-glucosylated glycoproteins.1, 7 We uncovered that calnexin expression increased upon tunicamycin treatment (Figures 1 and 3) and that the levels of calnexin expression inversely correlated with tunicamycin sensitivity (Figure 4). To determine whether the cytosolic domain of calnexin (with no demonstrated chaperone function) is involved in tunicamycin resistance, we aimed at expressing the C-terminal region of calnexin in MCF-7 cells to further reduce tunicamycin sensitivity. To this end, we generated a calnexin construct lacking most of the luminal domain (aa 53–422) but containing a signal peptide, a transmembrane domain and the entire cytosolic domain (ΔE, Figure 5a). The deletion of this domain completely abrogates the formation of the lectin-binding site as characterized by the structure of its luminal domain.12 MCF-7 cells were transfected with either the corresponding empty vector (Mock) or the vector containing this calnexin construct (ΔE). Polyclonal populations of cells resistant to G418 (400 μg/ml G418) were selected and the expression of calnexin ΔE was evaluated by immunoblot analysis. To ensure that the expression of ΔE calnexin in MCF-7 cells did not affect the ER stress response (due to an increased concentration of misfolded proteins in the ER or by affecting calcium homeostasis), we verified the expression of CHOP, GADD34 and EDEM mRNA using RT–PCR. These genes are the respective targets of the ER signalling kinases PERK and IRE1.13 The amount of CHOP, GADD34 and EDEM mRNA was comparable in MCF-7, MCF-7 Mock and MCF-7ΔE cells thus indicating no increased ER-stress signalling in MCF-7ΔE cells under basal conditions (data not shown). Lysates from MCF-7 (Mock) or ΔE cells were immunoprecipitated using CNX antibody (raised against the cytosolic domain) followed by immunoblotting using the same antibody. The apparent molecular weight of recombinant calnexin ΔE was approximately 35 kDa (Figure 5b). As the C-terminus of calnexin ΔE exhibits an acidic domain with a highly negative charge, the apparent molecular weight on SDS–PAGE is different from the calculated molecular weight (as for full-length calnexin). Cell fractionation studies confirmed that the ΔE protein was properly localized to the ER (data not shown). MCF-7 (Mock) and ΔE cells were treated or not with tunicamycin for 24 h and the amount of apoptotic cells, evaluated by TUNEL assay was compared to that obtained in MCF-7R and MCF-7+siRNA calnexin (S2). Surprisingly, MCF-7ΔE cells showed an increased sensitivity to tunicamycin comparable to that observed in S2 siRNA-transfected cells (Figure 5c). Indeed, more than 20% of the ΔE cells were apoptotic after 24 h of treatment (Figure 5c) as compared to 5% of the Mock cells. Interestingly, the expression of endogenous full-length calnexin still increased upon tunicamycin treatment in MCF-7ΔE cell line (Figure 5d). In addition, both cell lines (MCF-7 Mock and ΔE) expressed similar levels of full-length calnexin protein when normalized to the levels of the cytosolic kinase ERK-1 (Figure 5d). As ΔE cells were more sensitive to tunicamycin-induced apoptosis than Mock cells, we tested whether this effect was due to the association of calnexin with its cognate substrates and therefore related to a lack of effect of tunicamycin on oligosaccharyl transferase. To this end, MCF-7ΔE and Mock cells were treated with tunicamycin for 90 min, including a terminal 15 min pulse-label with 35S-methionine. In both cell types, tunicamycin treatment abrogated calnexin-nascent glycoprotein chains association (Figure 5e), thus suggesting that the observed sensitivity of MCF-7ΔE cells to tunicamycin-induced apoptosis may be dependent on calnexin cytosolic and transmembrane domains.
Construction and characterization of tunicamycin effect on a calnexin lacking the luminal domain. (a) The fragment corresponding to the amino acid 53–422 was removed from canine calnexin cDNA by restriction digestion. The construct generated encodes for a calnexin protein lacking amino acid 53–422 (ΔE). (b) MCF-7 cells were transfected by electroporation with pcDNA3 (Mock) or the same vector containing the calnexin protein lacking amino acid 53–422 (ΔE). Polyclonal cultures were selected using 400 μg/ml G418. The presence of ΔE was assessed by immunoprecipitation of Mock or ΔE cell lysates with anti-calnexin followed by immunoblotting with anti-calnexin (CNX) antibody. (c) MCF-7R, ΔE, Mock and S2 cells were treated for 24 h with 10 μg/ml tunicamycin. The number of apoptotic cells, determined by TUNEL assay, was expressed as a percentage of total input. The graph shows the mean of three independent experiments. (d) MCF-7 Mock and ΔE cells were incubated (+) or not (−) with 10 μg/ml tunicamycin for 24 h. Cells extracts were either directly immunoblotted with anti-ERK-1 and anti-calnexin (CNX) antibodies. Results are the representative of three independent experiments. (e) MCF-7 Mock and ΔE cells were incubated (+) or not (−) with 10 μg/ml tunicamycin for 75 min then pulse labeled with 35S-Methionine for additional 15 min. Cells were then lysed and calnexin immunoprecipitated. Immunoprecipitated material was then resolved by SDS-PAGE followed by fluorography analysis
Mechanisms of calnexin-mediated tunicamycin-induced apoptosis in MCF-7 cells
As MCF-7R expressed higher basal calnexin expression levels and since ΔE cells were more sensitive to tunicamycin-induced apoptosis than the resistant cell line (MCF-7R), we concluded that although total (endogenous and recombinant) calnexin expression was higher in MCF-7R and ΔE cells, tunicamycin effects were opposite. This led us to postulate that the association of calnexin with specific, nonglycan-linked partners may be altered upon tunicamycin treatment and may result in a phenotypic resistance to tunicamycin-induced apoptosis. Recently, Zuppini et al.10 reported that calnexin associates with the ER resident protein Bap31, a caspase-8 substrate, whose proteolytic cleavage results in apoptosis induction.14 We therefore, evaluated the cleavage of Bap31 in response to tunicamycin in our three cell lines MCF-7R, Mock or ΔE by immunoblot (Figure 6). Upon tunicamycin treatment, Bap31 was found to be cleaved only in ΔE cells, resulting in the production one fragment that is detected by immunoblot using anti-Bap31 antibodies (Bap20; Figure 6a). In parallel, to ensure that caspase-8, the protein responsible for Bap31 cleavage, was activated, we tested caspase-8 activity reflected by its autoproteolysis by immunoblot on protein extracts from cells treated (+) or not (−) with tunicamycin (Figure 6b). Caspase-8 was activated to a similar extend in response to tunicamycin in both Mock and ΔE cells (Figure 6b). This demonstrated that the apoptotic pathway initiated by caspase-8 was functional in both cell lines and suggested that the difference observed for Bap31 proteolysis may reside at the level of Bap31 itself. Interestingly, the basal association of Bap31 with calnexin was significantly higher (approximately × 3) in ΔE cells than in Mock cells (Figure 6c, right blot and Figure 6d), whereas the expression of Bap31 was similar in both MCF-7 Mock and ΔE cells (Figure 6c, left blot). To verify the presence of ΔE in association with Bap31, the reverse experiment was carried out by immunoprecipitating Bap31 followed by immunoblotting using CNX antibodies (Figure 6e). These results demonstrated that Bap31 immune complex contained calnexinΔE as well as the endogenous calnexin in MCF-7 ΔE cells, thus indicating that ΔE and Bap31 also form a complex and calnexin/Bap31 association may, therefore, occur at the level of calnexin transmembrane and/or cytosolic domain.
Caspase 8 activation upon tunicamycin treatment in MCF-7 cells. (a) MCF-7 Mock, ΔE and MCF-7R cells were treated with 10 μg/ml tunicamycin for 3 h. Cells extracts were directly immunoblotted with anti-Bap31 and anti-calnexin (CNX) antibodies. (b) MCF-7 Mock and ΔE cells were treated as described in a. The proteolytic cleavage of caspase 8 was assessed by immunoblot using anti-caspase 8 antibody. (c) MCF-7 Mock and ΔE cells extracts were either directly immunoblotted (WCL) or calnexin immunoprecipitated (Ip) prior immunoblot using the indicated antibodies. The amount of Bap31 associated with calnexin is reported as % of total Bap31 in panel (d). (e) MCF-7 Mock and ΔE cells extracts were either directly immunoblotted (WCL) or Bap31 immunoprecipitated (Ip) prior immunoblot using anti-calnexin (CNX) antibodies. Results are representative of three independent experiments
Our data show that although Bap31 expression was similar in MCF-7 Mock and ΔE cells (Figure 6c), its caspase-8 mediated cleavage in Bap20 fragment was generated only in ΔE cells (Figure 6a), which correlates with higher apoptosis sensitivity. This led us to postulate that calnexin cytosolic and/or transmembrane domains (ΔE) might serve as scaffold to allow Bap31 cleavage by caspase-8. To test this hypothesis, the association of Bap31 with calnexin was assessed upon tunicamycin treatment in both MCF-7 Mock and ΔE cells. Lysates from MCF-7 Mock or ΔE cells were collected after 0, 3 and 24 h of 10 μg/ml tunicamycin treatment, immunoprecipitated with anticalnexin antibodies and immunoblotted using anti-Bap31 and CNX antibodies. Our results indicated that, in Mock cells, Bap31/calnexin association was not affected significantly by a tunicamycin treatment (Figure 7a). In contrast, in MCF-7 ΔE cells, this complex was destabilized upon tunicamycin treatment with the observed dissociation/degradation of more than 50% of Bap31 during the first 3 h of treatment (Figure 7a and b). This pattern was dramatically altered when the experiment was performed in presence of the caspase-8 inhibitor Z-IETD-FMK (Figure 7c and d). Indeed, whereas in MCF-7 Mock cells, the treatment did not result in any major change on the kinetics of Bap31-calnexin association, it altered the association of Bap31 and calnexin in ΔE cells (Figure 7d), to mimic the association pattern observed in Mock cells (Figure 7b and d). This suggested that the dissociation of calnexin and Bap31 depends on caspase-8 activity and therefore confirms our initial hypothesis that calnexin cytosolic domain might serve as scaffold to allow Bap31 cleavage by caspase-8.
Calnexin-dependent apoptotic machinery in MCF-7 Mock and ΔE cells. (a) MCF-7 Mock and ΔE cells were with 10 μg/ml tunicamycin for the indicated periods of time. Lysates were then immunoprecipitated using anti-calnexin antibody, immunoprecipitates were then resolved by SDS-PAGE and subjected to immunoblotting with either anti-Bap31 or anti-calnexin (CNX) antibodies. (c), Same experiments were repeated in the presence of 40 μM of the caspase 8 inhibitor (Z-IETD-FMK). These experiments were repeated three times and quantified. Quantification±S.D. is shown in panels b and d respectively in the absence or presence of the caspase 8 inhibitor. Results are representative of four independent experiments
Discussion
In this study, we investigated the potential role of calnexin in tunicamycin-induced apoptosis (i.e. ER stress-induced). To this end, we used the well characterized human breast cancer cell line MCF-7 which is relatively resistant to apoptosis-induced by tunicamycin compared to other cell lines (Figure 1a). This resistance may be related either to the absence of caspase-3, a caspase activated in response to apoptotic stimuli,15 or to the deregulation of molecular chaperone expression.6 However, no demonstration of these hypotheses has been published to date. To directly investigate the possible contribution of caspase-3 deficiency in tunicamycin-induced apoptosis in MCF-7 cells, we measured tunicamycin-induced apoptosis levels in MCF-7 cells in which caspase-3 expression was reconstituted (MCF-7 Casp3) (Figure 2). Our data showed that reconstitution of caspase-3 expression had no effect on tunicamycin-induced apoptosis sensitivity. Interestingly, upon tunicamycin treatment, the expression levels of the chaperone calnexin strongly increased specifically in MCF-7 cells (Figure 1d and e). Recently, it has been shown that a caspase-mediated cleavage of the cytoplasmic domain of calnexin is involved in resistance to apoptosis.9 As a consequence, we tested whether calnexin cleavage may occur in MCF-7 Casp3 and MCF-7 cells in response to tunicamycin (Figure 2d). Our results indicated that the cytoplasmic tail of calnexin was not cleaved upon tunicamycin treatment and thus, did not act in resistance to tunicamycin-induced apoptosis in our experimental model. We then investigated whether MCF-7 cells resistant to ER stress-induced apotosis displayed altered calnexin expression levels. Interestingly, calnexin basal expression was two- to three folds higher in MCF-7R cells than in MCF-7 cells. In addition, tunicamycin treatment did not significantly affect calnexin expression in these cells, thus suggesting that elevated calnexin expression levels could protect cells from apoptosis as previously reported in other cell lines.16 To directly investigate the possible interference of calnexin with apoptotic programs, the induction apoptosis was measured in MCF-7 and MCF-7R cells in which calnexin expression had been silenced using siRNA (S2) (Figure 4). Our data show that depletion of calnexin sensitized cells to tunicamycin-induced apoptosis. Although calnexin expression was still increased upon tunicamycin treatment, siRNA-mediated reduction of its expression correlated with increased tunicamycin-induced apoptosis. We also examined whether the observed resistance to tunicamycin-induced apoptosis of MCF-7 cells was common for other pro-apoptotic agents (Figure 4f). MCF-7 cells were particularly resistant to treatments with tunicamycin, DTT, Genistein and Herbimicyn A but were sensitive to cell death induced by Azetidine-2-Carboxylic Acid (AZC). Interestingly, cells exhibiting a reduced calnexin expression displayed an enhanced sensitivity to tunicamycin and DTT specifically whereas no further effect was observed for AZC, Herbimicyn A and Genistein. Furthermore, MCF-7R cells which were selected based on their resistance to tunicamycin treatment, also displayed resistance to DTT treatments and a reduced sensitivity to AZC. The latter observation may be possibly explained by the fact that in MCF-7R cells, the ER folding machinery is preconditioned to handle a large amount of misfolded proteins. Therefore, the contribution of secretory protein unfolding to AZC-induced apoptosis may be reduced. Together, these data indicate that calnexin expression level may represent a good predictor of cell sensitivity to ER-mediated cell death.
Apoptosis induction by tunicamycin has been extensively documented in various cell types. For instance, treatment of SH-SY5Y neuroblastoma cells by tunicamycin leads to rapid changes in the expression of target genes of the Unfolded Protein Response. However, prolonged treatments also trigger a delayed caspase-dependent cell death.17 The molecular mechanisms by which these events are triggered are now better understood.17 Indeed, in many instances, programmed cell death involves an obligatory propagation through mitochondria, causing egress of critical proapoptotic regulators to the cytosol.17 These regulators include an antiapoptotic group comprising BCL-2 and BCL family members and a proapoptotic group with BAK, BAX. The balance between these opposing members is essential and is regulated by a third group called the BH3-domain-only proteins,17 which promote the mitochondrial phase of apoptosis. In addition, this phase requires a large-scale fission step that is dependent on the dynamin-like GTPase Drp117 and is triggered by the caspase-8-dependent cleavage of the ER-resident integral membrane protein Bap31.17 Finally, a recent report by Zuppini et al. 10 reported that calnexin and Bap31 formed a complex and suggested their participation in ER-stress-mediated apoptosis. Although our data are globally in accordance with this observation, the differences observed in both studies can be explained by the fact that Zuppini et al. used NKR cells which may have adapted to the absence of calnexin by compensatory mechanisms.
Inhibition of N-glycosylation by tunicamycin was shown to prevent the association of nascent glycoprotein chains with calnexin, but calnexin's chaperone function resides in its luminal domain. To determine if the cytosolic domain of calnexin was involved in tunicamycin resistance, we expressed in MCF-7 cells, a calnexin construct consisting of a short, nonfunctional portion of the luminal domain12 as well as the transmembrane and cytosolic domains (ΔE). MCF-7 cells expressing this truncated protein (MCF-7 ΔE) exhibited a restored sensitivity to tunicamycin-induced apoptosis. We, therefore, tested whether the stress-mediated apoptosis correlated with the association of calnexin with molecules previously reported to be involved in proapoptotic mechanisms. Whereas the association of calnexin BAK, previously reported in S. pombe,18 was not detected in our experimental system (data not shown), a strong association of calnexin with the ER integral membrane protein Bap31 was found only in MCF-7 ΔE cells in response to tunicamycin (Figure 6). Surprisingly, under basal condition the abundance of Bap31/calnexin complex was higher in MCF-7 ΔE cells than in MCF-7 Mock cells (15 vs >5%; Figure 6c and d). This observation most likely results from the interaction of the calnexin construct lacking most of the luminal domain with Bap31 (Figure 6e). When MCF-7 Mock and ΔE cells were treated with tunicamycin, the association of Bap31 and calnexin remained unchanged in MCF-7 cells whereas it decreased in cells stably expressing ΔE (Figure 7a and b), suggesting that the regulation of this association is involved in the control the tunicamycin-induced apoptosis.
Moreover, we found that caspase-8, which is responsible for the cleavage of Bap31 (14), was activated in a similar manner in both MCF-7 Mock and ΔE cells upon tunicamycin treatment (Figure 6b). The treatment of these cells with the caspase-8 inhibitor (Z-IEDT-FMK) stabilized the complex formed by calnexin and Bap31 in ΔE cells (Figure 7c and d). Therefore, this shows that calnexin-Bap31 dissociation is dependent on caspase-8 activity. Interestingly, upon tunicamycin treatment, Bap31 dissociated from calnexin in MCF-7 ΔE cells, thus providing a correlation between calnexin and caspase-8 cleavage for Bap31. These results led us to postulate that calnexin/Bap31 association might represent a docking complex allowing caspase-8 cleavage of Bap31 as previously suggested by Ng et al. 14 It is thus conceivable that the cytoplasmic tail of calnexin may mediate the interaction between Bap31 and calnexin.
In summary, we have demonstrated that the levels of calnexin upon tunicamycin treatment correlate with cell's sensitivity to this drug. We were able to restore partially MCF-7 sensitivity to tunicamycin-induced apoptosis either by reducing calnexin expression levels or by expressing a calnexin construct lacking the ER luminal domain. These findings led us to propose a novel mechanism for the control of apoptosis where the cytoplasmic domain of the ER chaperone calnexin contributes to ER stress-induced apoptosis by acting as a scaffold that promotes Bap31 cytosolic domain cleavage by caspase-8, and thus generating the proapoptotic p20 fragment. Thus, utilizing the properties of MCF-7 cell line to resist to tunicamycin-induced apoptosis, we have characterized a molecular mechanism by which calnexin regulates ER-stress-mediated pathways in a manner independent of its chaperone functions but dependent on its association with Bap31.
Materials and Methods
Antibodies, proteins and constructs
Anti-calnexin antibodies (Stressgen, Victoria, BC Canada) was used for immunoblotting and immunoprecipitations as previously described.11 Anti-Bap31 antibody was described previously.14 Anticaspase-8 antibody was purchased from Upstate Biotechnology and recognized both isoform caspase-8a (55 kDa) and b (54 kDa) and their 20 kDa proteolytic fragment. Calnexin lacking amino acids 53–422 (ΔE) was constructed by digestion with Eco47III restriction enzyme from canine calnexin cDNA followed by ligation and subcloning into pcDNA3 eukaryotic expression vector (Invitrogen, CA, USA). Bacterial recombinant carboxy terminal domain of calnexin was obtained by thrombin cleavage of GST-calnexin cytosolic domain on glutathione-Sepharose beads, followed by benzamidine-agarose chromatography of the supernatant and dialyze-concentration. The caspase-8 Inhibitor Z-IETD-FMK was from Calbiochem and used at the final concentration of 40 ΔM as previously described.19
Cell culture and transfection
Rat2, COS7, MDCK, Jurkat, HuH7, Hela, HepG2 and MCF-7 cells were passaged every three days at 3 × 103 cells/cm2 in Dulbecco's modified eagle medium (DMEM) containing 10% FBS at 37°C, 5% CO2. MCF-7 cells were electroporated with a Bio-Rad Gene-Pulser at 260 V and 960 μF with either an empty vector pcDNA3 (Mock) or with the same vector containing the ΔE construct (MCF-7ΔE). Polyclonal cultures were selected in DMEM containing 10% FBS and 400 μg/ml G-418. MCF-7 Casp3 cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) with the vector CMV-Myc-Caspase3 containing the caspase-3 sequence, according to the protocol supplied by the manufacturer (Ambion Inc., Austin, TX, USA).
Protein identification by mass spectrometry
Calnexin was identified as described previously.20 Briefly, the relevant band was excised from the polyacrylamide gel, reduced, alkylated using iodoacetamide, and digested with trypsin. Extracted peptides were applied onto a reverse phase guard column and then eluted in-line on a 10 cm by 75 μm PicoFrit column filled with BioBasic C18. Samples were eluted with a linear gradient of 5–70% acetonitrile/0.1% formic acid. A 2000-V charge was applied to the PicoFrit column such that the eluted peptides were electrosprayed into a Cap liquid chromatography quadrupole time-of-flight (LC-qToF) mass spectrometer (MicroMass). Peptides were analyzed on MS/MS mode and results analyzed using the Mascot Software (Matrix Science).
Immunoprecipitation and immunoblot
Cells were solubilized for 30 min on ice in buffer A (30 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM PMSF, 1 mM NaF, 1 μg/ml leupeptin, 5 kU/ml aprotinin containing 1.5% CHAPS). The lysate was clarified by centrifugation at 435 000 × gmax for 30 min at 4°C and the supernatant was incubated overnight at 4°C with the indicated antibody. Protein A Sepharose beads were added for 1 h at 4°C with rotation then pelleted and then washed four times with buffer A. Beads were resuspended in Laemmli sample buffer.21
Metabolic labeling
Cells were maintained in DMEM supplemented with 10% FBS, 20 mM HEPES, pH 7.6 and 1% penicillin/streptomycin (complete DMEM). For metabolic labeling, experiments near confluent cultures in 55-mm dishes were washed in DMEM (minus methionine or minus phosphate) containing 10% dialyzed FBS and preincubated for 60 min in the same media. Cells were then pulsed either for 15 min with 100 μCi/ml of 35S-label. At the end of the labeling period, dishes were immediately placed on ice. The cells were washed twice with ice-cold PBS (50 mM phosphate, pH 7.5/150 mM NaCl) and once with ice-cold HBS (50 mM HEPES, pH 7.6/200 mM NaCl). Cells were collected and lysed for 30 min on ice in buffer A as described above.
Development of the tunicamycin-resistant cell line
The MCF-7 cell line resistant to tunicamycin (MCF-7R) was developed in our laboratory as illustrated in Figure 2a. The MCF-7R cell line was cultivated under similar conditions as the MCF-7 cells except with addition of 10 μg/ml tunicamycin to the growth medium. MCF-7 cells were plated in a 24-well plate (2 × 104 per well). After 2 days (0 m), the medium was replaced with fresh medium containing 10 μg/ml tunicamycin and the medium was renewed every day until day 3 for the first 2 weeks and then once a week. After 1 month (1 m), colonies of growing cells appeared. After a total of 4 months (4 m), three colonies were transferred to individual T25 flask and subculture was performed during the next 6 months. During this period, MCF-7R cells had a slow, but increasing proliferation rate, which reached a weekly split ratio of about 1 : 10 between month 10 and 12, where after the cells were routinely subcultivated with a weekly split ratio of about 1 : 20.
siRNA transfection
The human calnexin siRNA (CANX_2 siRNA and CANX_3 siRNA), nonsilencing siRNA (Silencer Negative Control #1 siRNA) and transfection agent (siPORT Lipid) were obtained from Ambion. MCF-7 cells were transfected with siRNA using siPORT Lipid according to the manufacturer's protocol.
Evaluation of the rate of apoptosis
Cells were treated for the indicated periods of time with 10 μg/ml tunicamycin. Floating cells were collected by centrifugation and attached cells were trypsinized prior to centrifugation. Both cell populations were then mixed, washed in cold PBS and fixed in 100% methanol. Apoptosis rates were then evaluated by measuring caspase activity with a CytoDeath™ mAb that recognizes cleaved cytokeratin 18 (Boehringer-Mannheim). Alternatively, DNA strand breaks were detected using a TUNEL assay as described previously,22 we used the ApoBrdU kit and MPlus software (Phoenix Flow Systems), this was reported to the proportion of cells with sub-G1 DNA levels with propidium iodide.
MTT assay
For proliferation assay, cells were plated in triplicates in 96-well dishes, at 5000 cells/100 μl in 2% FCS. Cells were stimulated or not with apoptosis-inducing agents and grown over a 24 h period. Cell proliferation was assessed using the non-radioactive MTT cell proliferation assay for eukaryotic cells (Cell Titer 96, Promega G 4000). Absorbance was measured at 570 nm with a reference wavelength at 450 nm, using a Bio-tek Microplate reader.
Abbreviations
- ER:
-
endoplasmic reticulum
- siRNA:
-
small interfering RNA
- SDS-PAGE:
-
sodium dodecyl sulfate polyacrylamide gel electrophoresis
References
Zapun A, Jakob CA, Thomas DY, Bergeron JJ . Protein folding in a specialized compartment: the endoplasmic reticulum. Structure Fold Des 1999; 7 (8): R173–R182.
Chevet E, Cameron PH, Pelletier MF, Thomas DY, Bergeron JJ . The endoplasmic reticulum: integration of protein folding, quality control, signaling and degradation. Curr Opin Struct Biol 2001; 11 (1): 120–124.
Rao RV, Bredesen DE . Misfolded proteins, endoplasmic reticulum stress and neurodegeneration. Curr Opin Cell Biol 2004; 16 (6): 653–662.
Kaufman RJ . Orchestrating the unfolded protein response in health and disease. J Clin Invest 2002; 110 (10): 1389–1398.
Parodi AJ . Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem J 2000; 348 (Part 1): 1–13.
Gazit G, Lu J, Lee AS . De-regulation of GRP stress protein expression in human breast cancer cell lines. Breast Cancer Res Treat 1999; 54 (2): 135–146.
Bergeron JJ, Brenner MB, Thomas DY, Williams DB . Calnexin: a membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem Sci 1994; 19 (3): 124–128.
Chevet e.Calnexin . Alliance for Cellular Signaling. AfCS-Nature Molecule Pages 2004.
Takizawa T, Tatematsu C, Watanabe K, Kato K, Nakanishi Y . Cleavage of calnexin caused by apoptotic stimuli: implication for the regulation of apoptosis. J Biochem (Tokyo) 2004; 136 (3): 399–405.
Zuppini A, Groenendyk J, Cormack LA, Shore G, Opas M, Bleackley RC et al. Calnexin deficiency and endoplasmic reticulum stress-induced apoptosis. Biochemistry 2002; 41 (8): 2850–2858.
Chevet E, Wong HN, Gerber D, Cochet C, Fazel A, Cameron PH et al. Phosphorylation by CK2 and MAPK enhances calnexin association with ribosomes. EMBO J 1999; 18 (13): 3655–3666.
Schrag JD, Bergeron JJ, Li Y, Borisova S, Hahn M, Thomas DY et al. The Structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol Cell 2001; 8 (3): 633–644.
Schroder M, Kaufman RJ . The mammalian unfolded protein response. Annu Rev Biochem 2005; 74: 739–789.
Ng FW, Nguyen M, Kwan T, Branton PE, Nicholson DW, Cromlish JA et al. p28 Bap31, a Bcl-2/Bcl-XL- and procaspase-8-associated protein in the endoplasmic reticulum. J Cell Biol 1997; 139 (2): 327–338.
Turner CP, Blackburn MR, Rivkees SA . A1 adenosine receptors mediate hypoglycemia-induced neuronal injury. J Mol Endocrinol 2004; 32 (1): 129–144.
Tomassini B, Malisan F, Franchi L, Nicolo C, Calvo GB, Saito T et al. Calnexin suppresses GD3 synthase-induced apoptosis. FASEB J 2004; 18 (13): 1553–1555.
Breckenridge DG, Germain M, Mathai JP, Nguyen M, Shore GC . Regulation of apoptosis by endoplasmic reticulum pathways. Oncogene 2003; 22 (53): 8608–8618.
Torgler CN, de Tiani M, Raven T, Aubry JP, Brown R, Meldrum E . Expression of bak in S. pombe results in a lethality mediated through interaction with the calnexin homologue Cnx1. Cell Death Differ 1997; 4 (4): 263–271.
Pirnia F, Schneider E, Betticher DC, Borner MM . Mitomycin C induces apoptosis and caspase-8 and -9 processing through a caspase-3 and Fas-independent pathway. Cell Death Differ 2002; 9 (9): 905–914.
Wong HN, Ward MA, Bell AW, Chevet E, Bains S, Blackstock WP et al. Conserved in vivo phosphorylation of calnexin at casein kinase II sites as well as a protein kinase C/proline-directed kinase site. J Biol Chem 1998; 273 (27): 17227–17235.
Laemmli UK . Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227 (5259): 680–685.
Nantel A, Mohammad-Ali K, Sherk J, Posner BI, Thomas DY . Interaction of the Grb10 adapter protein with the Raf1 and MEK1 kinases. J Biol Chem 1998; 273 (17): 10475–10484.
Acknowledgements
We thank the Chevet's lab and D Fessart for critical reading of the manuscript. This work was supported by a grant from the Canadian Institutes for Health Research to EC. FD was supported in part by an operating grant from Genome Quebec/Canada to the Montreal Proteomics Network – the cell map project. EC is a junior scholar from the FRSQ. AN was supported by the NRC Genome Health Initiative. This is NRC publication number 46182.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by SJ Martin
Rights and permissions
About this article
Cite this article
Delom, F., Emadali, A., Cocolakis, E. et al. Calnexin-dependent regulation of tunicamycin-induced apoptosis in breast carcinoma MCF-7 cells. Cell Death Differ 14, 586–596 (2007). https://doi.org/10.1038/sj.cdd.4402012
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4402012
Keywords
This article is cited by
-
Klotho modulates ER-mediated signaling crosstalk between prosurvival autophagy and apoptotic cell death during LPS challenge
Apoptosis (2019)
-
Calnexin, an ER-induced protein, is a prognostic marker and potential therapeutic target in colorectal cancer
Journal of Translational Medicine (2016)
-
The ATF6 branch of unfolded protein response and apoptosis are activated to promote African swine fever virus infection
Cell Death & Disease (2012)
-
Multiphoton ANS fluorescence microscopy as an in vivo sensor for protein misfolding stress
Cell Stress and Chaperones (2011)
-
BAP31 is frequently overexpressed in patients with primary colorectal cancer and correlates with better prognosis
Chinese Science Bulletin (2011)