Abstract
Formation of the CD95 (APO-1/Fas) death inducing signaling complex (DISC) plays a central role in CD95 signaling. Previously, CD95 DISC composition was analyzed by two-dimensional gel electrophoresis and four major cytotoxicity-associated proteins (CAP1–4) were found. CAP1 and CAP2 were defined to be unmodified and phosphorylated FADD, respectively. CAP4 was identified as procaspase-8a. CAP3, however, has remained elusive. In this study, we demonstrate that CAP3 is an intermediate of procaspase-8 processing. CAP3 is generated within seconds of DISC formation and subsequently processed to the prodomain of procaspase-8a that is known as p26 (CAP5). These findings lead to new insights into the mechanism of procaspase-8 processing and apoptosis initiation.
Similar content being viewed by others
Introduction
CD95 (Fas/APO-1) is a member of the death receptor family, a subfamily of the TNF-R superfamily.1, 2, 3, 4, 5 Death receptors contain an intracellular death domain (DD) that plays a central role in the transduction of the apoptotic signal. Crosslinking of CD95 with its natural ligand CD95L (CD178)6 or with agonistic antibodies such as anti-APO-17 induces apoptosis in sensitive cells. A death inducing signaling complex (DISC) is formed upon CD95 stimulation within seconds.8 The DISC consists of oligomerized CD95 molecules, the adaptor fas-associated DED (FADD), procaspase-8, procaspase-10 and c-FLICE inhibitory protein (c-FLIP)L/S/R.5, 9 The stoichiometry of the DISC components, however, has not been unambiguously established yet. The contacts between the molecules in the DISC are based on homophilic interactions. The DD of CD95 interacts with the DD of FADD while the death effector domain (DED) of FADD interacts with the N-terminal tandem DED of procaspase-8.
Two isoforms of procaspase-8 (procaspase-8a and -8b) were reported to be bound to the DISC.10 Both isoforms possess two tandem DED domains as well as the catalytical subunits p18 and p10. Procaspase-8a contains an additional 2 kDa (15 amino-acid (aa)) fragment, which results from the translation of exon 9. This small fragment is located between the second DED and the large catalytical subunit resulting in different length of the procaspases-8a and -8b, p55 and p53, respectively.
Binding of procaspases-8a and -b to the DISC results in their autocatalytic cleavage for which a two-step mechanism has been described.11 The initial cleavage at aspartate (Asp)374 generates the two subunits p43/p41 and p12. In a second step cleavage takes place at Asp216 and Asp384, producing the active enzyme subunits p18, p10 and the prodomains p26/p24. As a result of procaspase-8 processing the caspase-8 heterotetramer (p18/p10)2 is formed at the DISC, which is subsequently released into the cytosol starting the apoptotic signaling cascade.12
In the initial analysis of the CD95 DISC using two-dimensional (2-D) gels four major differential protein spots were revealed.8 Those four spots were considered as four proteins recruited to CD95 and being responsible for the cytotoxicity induced by anti-APO-1. Thus, they were called cytotoxicity-associated proteins (CAPs). CAP1 was shown to be FADD, and CAP2 turned out to be a phosphorylated form of FADD. CAP4 was originally sequenced by nanoelectrospray tandem mass spectrometry and identified as procaspase-8a.13 Subsequently, when the composition of the CD95 DISC was examined under prolonged stimulation two other CAPs (CAP5 and CAP6) were discovered.11 CAP5 and CAP6 were shown to be the prodomains of procaspase-8a and -b, respectively. CAP3, however, has remained elusive.
Electrospray ionization-tandem mass spectrometry (ESI-MS/MS) analysis of CAP3 revealed the identity of two peptides to the procaspase-8 N-terminus.13 Based on this finding there have been several hypotheses referring to the identity and function of CAP3: CAP3 might be a procaspase-8 prodomain13 or a short splice variant of procaspase-8 like c-FLIPS or c-FLIPL playing an inhibitory role in CD95 signaling.14 Alternatively, CAP3 might be an inducible cofactor required for procaspase-8 activation.11, 15 In this paper, we show decisive experiments demonstrating that CAP3 is an intermediate cleavage product of procaspase-8a generated at the DISC. We demonstrate that CAP3 is the N-terminal part of procaspase-8a (aa 1–216). Cleavage of p43 at Asp216 results in the formation of the large subunit p18 and CAP3, which, in turn, is processed to p26 (aa 1–210). These results provide a new understanding of the mechanism of procaspase-8 activation at the DISC.
Results
CAP3 is present at the CD95 DISC of different cell lines
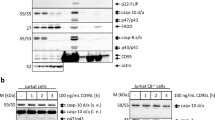
In previous studies CAP3 was found by 2-D gel analysis of the CD95 DISC.8, 11 It was detected either by autoradiography, when the DISC was immunoprecipitated from metabolically labeled cells with agonistic anti-APO-1 antibodies, or by silver staining followed by nanoelectrospray tandem mass spectrometry analysis. Initially, we aimed to detect CAP3 at the DISC by a 2-D gel approach in combination with immunoblotting. With the specific anticaspase-8 monoclonal antibody (mAb) N2 that recognizes the N-terminus of procaspase-8, we detected the CAP3 spot at the DISC from SKW6.4 B lymphoblastoid cells. All other reported CAP proteins of the CD95 DISC were also present, namely FADD (CAP1), phosphorylated FADD (CAP2), procaspase-8a (CAP4), p26 (CAP5) and p24 (CAP6) resulting in a typical pattern (Figure 1a). The positions of the CAP spots were in agreement with their reported molecular masses and isoelectric points (pIs): CAP1 (27 kDa, pI 5.2), CAP2 (28 kDa, pI 5.15), CAP3 (27 kDa, pI 5.05), CAP4a (55 kDa, pI 4.9), CAP4b (53 kDa, pI 5.0), CAP5 (26 kDa, pI 4.75) and CAP6 (24 kDa, pI 4.86).8, 11 The spot corresponding to procaspase-8 (CAP4), was not resolved in an optimal way due to the signal from the heavy chain of the immunoprecipitating antibody which shows almost the same pI and molecular mass as procaspase-8.
CAP3 is present at the CD95 DISC of different cell lines. (a). The CD95 DISC was immunoprecipitated from 108 SKW6.4 cells stimulated with anti-APO-1 for 5 min. Immunoprecipitations were performed using anti-APO-1 in combination with Protein-A Sepharose. Immunoprecipitates were subjected to 2-D gels and immunoblotted with anticaspase-8 mAb N2 and anti-FADD mAb. Migration positions of full-length procaspase-8 (p55/p53), and its further processed products p43/p41 and p26/24 are indicated both in 2-D and 1-D gels. The positions of CAP1 (FADD), CAP2 (phosphorylated FADD), CAP3, CAP4 (procaspase-8), CAP5 (p26), CAP6 (p24) are indicated by arrows. (b) Cells (108) each of H9, CEM, JA3 and JA3-Caspase-8 deficient were stimulated with anti-APO-1 for 5 min. Subsequently, CD95 DISCs were immunoprecipitated with Protein-A Sepharose and analyzed on 2-D gels by immunoblotting using the anticaspase-8 mAb N2. The positions of CAP3 and p26/p24 are indicated in the left panel. (c). Cells (108) each of indicated cell line were stimulated with anti-APO-1 for 5 min. Subsequently, CD95 DISCs were immunoprecipitated with Protein-A Sepharose and analyzed on 1-D 15% Anderson gels by immunoblotting using the anticaspase-8 mAb N2
To address the question whether CAP3 is present at the DISC of other cell lines, we examined DISCs immunoprecipitated from several T- and B-cell lines (Figure 1b). CAP3 was detected in the DISCs of both Type I (H9) and Type II (JA3, CEM) cells.11 The 2-D pattern of the CAP spots was in accordance with previous publications even though we applied a different method of detection. We did not observe CAP3 in the DISC of caspase-8-deficient cells. Thus, using 2-D gels in combination with immunoblotting, we observed CAP3 along with the other CAP spots at the DISC of several cell lines.
To simplify the experiments, we developed an approach for the detection of CAP3 by one-dimensional (1-D) gels with subsequent immunoblotting. The molecular mass of CAP3 is only slightly higher than the one of p26. Therefore, they are often indistinguishable in Laemmli gels. To enable a better separation of the proteins within the molecular mass range between 20 and 30 kDa, we used 15% Anderson gels.16 Thereby, we obtained a clear separation of CAP3 from p26 at the DISCs immunoprecipitated from several cell lines (Figure 1c). Molecular masses of bands that were detected with the anticaspase-8 mAb N2 were in accordance with those of CAP3, p26 (CAP5) and p24 (CAP6). Thus, we observed CAP3 at the DISC of several cell lines using both 1-D as well as 2-D gels.
Identification of CAP3 as the prolonged prodomain of procaspase-8a
To further characterize CAP3, we examined if CAP3 might be processing product of procaspase-8. HUT78 cells were stimulated with leucine zipper-CD95 ligand (LZ-CD95L) that was followed with immunoprecipitation using anticaspase-8 N1 mAb. N1 mAb recognizes the N-terminus of procaspase-8. CAP3 was detected only upon CD95 stimulation and procaspase-8 processing (Figure 2a). These results provided a first hint that CAP3 might be a procaspase-8 processing product.
CAP3 is the prolonged prodomain of procaspase-8. (a) Procaspase-8 was immunoprecipitated from 108 HUT78 cells untreated (−) or treated (+) with LZ-CD95L for five minutes. Immunoprecipitates were applied to 15% Anderson gels and immunoblotted with either anticaspase-8 mAb N2 (top) or anti-caspase-8 mAb C15 (bottom). Migration positions of CAP3, p26, p24 and p18 are indicated. (b) Scheme of procaspases-8a, -b and putative procaspase-8 prodomains used to identify CAP3: death effector domain (DED), D, Aspartic acid residues at the cleavage sites. The epitopes at the N-terminus of procaspase-8 recognized by the anticaspase-8 monoclonal antibodies N1, N2 and C15 are indicated. The exon missing in procaspase-8b is shown as white box encoding 15 aa. (c) Procaspase-8 processing products at the DISC from SKW6.4 cells (lane 1), in vitro translated prodomains 1–223 (lane 2), 1–216 (lane 3), 1–210 (lane 4) were subjected to 15% Anderson gels and analyzed by immunoblotting using anticaspase-8 mAb N2. As a positive control, SKW6.4 cells were stimulated for 5 min with anti-APO-1. Subsequently, the DISC was immunoprecipitated and procaspase-8 processing products were analyzed by immunoblotting using anti-caspase-8 mAb N2 (lane 1)
Three aspartates are located at the N-terminal site of the large catalytic subunit p18 and, thus, are possible cleavage sites that might result in CAP3 and p26 in the course of procaspase-8 processing. These sites are TISD210, REQD216 and QTLD223 with REQD216 being conserved between mouse and man.17 It is believed that the prodomains p26 (CAP5) and p24 (CAP6) are generated by cleavage of p43 and p41, at this site, respectively. To test the possibility that CAP3 is also a prodomain of procaspase-8, different putative prodomains were generated (Figure 2b), in vitro translated and analyzed by 15% Anderson gels. As shown in Figure 2c the construct comprising aa 1–216 of procaspase-8a has an electrophoretic mobility identical to CAP3. Furthermore, the construct consisting of aa 1–210 migrates as p26 (CAP5), suggesting that the p26 prodomain is generated by the cleavage at the Asp210 and that CAP3 is a longer prodomain comprising aa 1–216. These results suggest that CAP3 is a cleavage product of procaspase-8a at the DISC generated by cleavage at Asp216.
Interestingly, we also observe a band between p26 and p24 (Figure 2a and Figure 2c). The molecular mass of this product suggests that it might correspond to CAP3b, the prolonged prodomain of procaspase-8b. Thus, possible cleavage sites of procaspase-8b that would result in generation of CAP3b and p24 are Asp201 and Asp195, respectively (Figure 2b). It should be noted that detection of CAP3b is complicated as the signal from this band overlaps with the signal from p26.
Kinetics of CAP3 generation and processing at the DISC
The DISC is formed within seconds after receptor engagement.8 As we have already shown, CAP3 was detectable at the DISC within a short time after stimulation (Figure 1a, b). To exactly determine the kinetics of CAP3 appearance in the DISC, SKW6.4 cells were stimulated with anti-APO-1 for different periods of time. Subsequently, DISCs were immunoprecipitated (Figure 3a). Our data demonstrates that CAP3 was detectable at the CD95 DISC within the first few minutes after stimulation. After 30 min the intensity of the spots corresponding to CAP3 started to decrease, and after 60 min the intensity of spots was markedly reduced. In contrast, p26 (CAP5) and p24 (CAP6) were detectable after 5 min of stimulation and peaked at 10–30 min. Both proteins were still detectable after as long as 120 min of stimulation. Thus, the generation of CAP3 and CAP5/6 shows different kinetics.
The time course of CAP3 generation at the DISC. (a) In all, 108 SKW6.4 cells were stimulated with anti-APO-1 For the indicated periods of time. DISCs were immunoprecipitated and analyzed using 2-D gels with subsequent immunoblotting. The membranes were developed using the same exposure time. (b) In all, 108 HUT78 cells were stimulated with anti-APO-1 For the indicated periods of time. DISCs were immunoprecipitated and analyzed using 1-D 15% SDS-PAGE gels with subsequent immunoblotting. (c) In all, 108 SKW6.4 cells were stimulated with anti-APO-1 For the indicated periods of time. Procaspase-8 was immunoprecipitated as described in Figure 2a and its processing products were analyzed with 15% Anderson gels
To confirm this result by an independent approach, we analyzed the kinetics of CAP3 and p26/p24 generation at the DISC using 1-D gels (Figure 3b). Interestingly, using 1-D gels, we again observed that CAP3 peaked earlier than p26/p24.
To examine the total cellular caspase-8 pool, we stimulated SKW6.4 cells with anti-APO-1 for different times and immunoprecipitated procaspase-8 with the anticaspase-8 mAb N1 (Figure 3c). The time-dependent decrease of CAP3 in the lysates was taking place in a manner similar to that of procaspase-8 (p55/p53). After 60 min CAP3 was hardly detectable, whereas at that time point the amount of the prodomains p26 and p24 reached maximum levels (Figure 3c). These results suggest that CAP3 might be an intermediate product of processing of procaspase-8 that is quickly degraded.
In addition, we tried to examine the kinetics of procaspase-8 processing at the DISC within shorter periods of time, namely within seconds after the stimulation. However, the pattern of procaspase-8 processing for these time points was similar to the earliest time point shown in Figure 3 (data not shown; see also Scaffidi et al.10). Thus, it seems that initial processing of procaspase-8 occurs too fast to detect the initial cleavage steps at the DISC using standard immunoprecipitation procedure.
CAP3 generation at the DISC in the presence of zVAD-fmk
To get more insights into the mechanism of CAP3 generation, we analyzed procaspase-8 processing in the presence of the pan-caspase inhibitor benzoyl-valine-alanine-aspartate-fluoromethylketone (zVAD-fmk). Interestingly, upon addition of zVAD-fmk we did not observe a complete inhibition of procaspase-8 processing, either at the DISC or in cell lysates (Figure 4a). Namely, p43/p41 were observed both in cell lysates and at the DISC, however, active caspase-8 was not detected in the cell lysates. Interestingly, in comparison to the p26/p24 prodomains, the amount of CAP3 was significantly enhanced at the DISC upon zVAD-fmk treatment (Figure 4a and b). This can be visualized again in 15% Anderson gels (Figure 4a) and in 2-D gels (Figure 4b). This effect seems to be dose-dependent as the amount of CAP3 at the DISC was enhanced upon the increase in zVAD-fmk concentrations (Figure 4b and c).
ZVAD-fmk enhances procaspase-8 processing at the DISC towards CAP3. (a) CD95 was immunoprecipitated from 108 SKW6.4 cells stimulated with anti-APO-1 for 5 min. Cells were pretreated with 20 μM of zVAD-fmk for 30 min before stimulation of CD95 with anti-APO-1. DISCs were separated in 15% Anderson gels and analyzed using the anticaspase-8 mAb N2 (upper panel), the anticaspase-8 mAb C15 (middle panel) and the anticaspase-8 mAb C5 (lower panel). Procaspase-8 processing in the lysates was also probed using the same panel of antibodies (b). SKW6.4 cells were pretreated with 1 μM, 20 μM of zVAD-fmk for 30 min or left untreated. Subsequently, DISCs were prepared as in (a) and resolved on 2-D gels. The generation of CAP3, p26 and p24 was analyzed by immunoblotting with the anticaspase-8 mAb N2. The membranes were developed in parallel using the same exposure time. (c) SKW6.4 cells were pretreated with the indicated concentrations of zVAD-fmk and procaspase-8 was immunoprecipitated as described in Figure 2a. Procaspase-8 processing at the DISC was analyzed by 15% Anderson gels and immunoblotting with the anticaspase-8 mAb N2. (d) DISC activity using Biotin-VAD-fmk. SKW6.4 cells were pretreated for 30 min with Biotin-VAD-fmk and then immunoprecipitated as described in (a). The immunoblotting was performed using the anti-caspase-8 mAb N2 (left panel) and Streptavidin-HRP (right panel). The positions of procaspase-8 and its cleavage products are indicated. (e) HUT78 cells were pretreated with 20 μM of zVAD-fmk for 30 min or left untreated. Subsequently, DISCs were prepared as in (a) and resolved on 1-D gels. C-FLIPL processing at the DISC was analyzed using antiFLIP mAb NF6. P43-FLIPL cleavage product is indicated. The presence of other DISC components: CD95 was analyzed using anti-CD95 rabbit polyclonal antibody C20, procaspase-8 (p55/p53) as well as cleavage product p18 were analyzed using anticaspase 8 mAb C15
Notably, the only spot enhanced upon zVAD-fmk addition on 2-D gels corresponds to CAP3 (Figure 4b). The same was observed in 1-D gels within the region of 24–30 kDa (Figure 4c). This result provides additional evidence that the CAP3 spot in 2-D gels is indeed identical to the prolonged prodomain of procaspase-8a (aa 1–216).
The analysis of CAP3 formation was also performed in the presence of the predicted optimal caspase-8 inhibitor benzoyl-isoleucine-glutamate-threonine-aspartate-fluoromethylketone (zIETD-fmk). The action of zIETD-fmk was identical to the action of zVAD-fmk and resulted in enhancement of CAP3 at the DISC (data not shown).
To examine the mechanism of zVAD-fmk action at the DISC leading to CAP3 accumulation we assayed DISC activity with biotinylated active site-directed pan-caspase inhibitor (Biotin-zVAD-fmk) (Figure 4d). We observed labeling of partially processed caspase-8 (p43/41) as well as the fully processed large catalytical subunit of caspase-8 (p18). Biotin-VAD-fmk was not covalently bound to procaspase-8 (p55/53). Interestingly, we observed the modification of additional bands above p43/p41, that probably corresponds to the modification of cleavage products of procaspase-10, another DED-containing caspase present at the DISC. Thus, we have shown that zVAD-fmk blocks the components of the active heterotetramer of caspase-8 that might have a composition of p43/p18/p102, p432/p102 or fully processed of p102/p182 in accordance with previous publications.12, 18
The observation that Biotin-VAD-fmk does not bind to procaspase-8 (p55/p53) molecules bound to the DISC might suggest that this compound does not bind to the active center of procaspase-8 at the DISC, possibly due to spatial nonaccessibility. Therefore, it is reasonable to assume that also zVAD-fmk does not block caspase-8 activity at the DISC completely as a result of possible spatial nonaccessibility. This might explain why p43/p41 and CAP3 are generated upon zVAD-fmk addition. To examine if caspase-8 activity can be detected at the DISC in the presence of zVAD-fmk, we analyzed processing of c-FLIPL. C-FLIPL is known to be cleaved by caspase-8 at the DISC.19 The analysis of the CD95 DISC, that was immunoprecipitated from SKW6.4 cells in the presence or absence of zVAD-fmk, has demonstrated that cleavage of c-FLIPL to p43-FLIPL is not altered upon addition of zVAD-fmk (Figure 4e). Thus, we can conclude that zVAD-fmk does not completely block caspase-8 activity at the DISC explaining the generation of CAP3 upon zVAD-fmk addition.
CAP3 is generated from procaspase-8a at the DISC
To demonstrate that procaspase-8a is indeed processed to CAP3 at the DISC, several procaspase-8a mutants were generated. The Asp residues involved in the prodomain formation (Asp210 and Asp216) were substituted by alanines and three mutants were produced (D210A, D216A, D210A/D216A) (Figure 5a). Their processing was investigated in an in vitro caspase-8 cleavage assay (Figure 5b). In this assay the DISC isolated from SKW6.4 cells was incubated with in vitro translated, [35S]-labeled wild type procaspase-8a and the respective mutants. The [35S]-labeled in vitro translated procaspase-8a proteins are recruited to the DISC and cleaved. Accordingly, for wild-type procaspase-8a we observed the generation of p43 and p26 (Figure 5b, lane 1). The D216A mutant showed the same cleavage pattern as wild-type procaspase-8a (Figure 5b, lane 3). In contrast, generation of p26 was abrogated in the D210A and D210A/D216A mutants (Figure 5b, lanes 2, 4). However, processing of the mutant D210A led to an additional cleavage product the molecular mass of which is in accordance with that of CAP3 (Figure 5b, lane 2). This product is not formed by processing of the mutants D216A and D210A/D216A demonstrating that it indeed results from the cleavage at Asp216 (Figure 5b, lanes 3, 4). These experiments with procaspase-8a mutants clearly demonstrate that CAP3 is an intermediate product of procaspase-8a processing.
CAP3 is generated from procaspase-8a at the DISC. (a) Scheme of cloned procaspase-8a mutants (wilde-type, D210A, D216A, D210A/D216A). (b) In vitro caspase-8 cleavage assays with [35S]-labeled procaspase-8a mutants (wt, D210A, D216A and D210A/D216A) were performed at 4°C overnight. The products of processing (p55, p43, CAP3 and p26) are indicated by arrows. (c) In vitro caspase-8 cleavage assays with D210A-procaspase-8 and wild-type procaspase-8 were performed for indicated periods of time at room temperature (RT). zVAD-fmk was added at a final concentration of 50 μM
It should be noted that neither in our experiments nor in the previous work CAP3 has been clearly detected by procaspase-8a processing using this assay.10, 11, 20 In order to detect CAP3 we examined different conditions of the in vitro caspase-8 cleavage assay for wild-type procaspase-8a (Figure 5c). To this end, [35S]-labeled in vitro translated wild-type procaspase-8a was incubated for short time periods (1–120 min) with immunoprecipitated DISC. However, we did not detect CAP3 under these conditions. This was also the case when the assay was performed in the presence of zVAD-fmk (Figure 5c). Apparently, the substrate concentrations in the in vitro caspase-8 cleavage assay are different from those at the DISC in vivo resulting in different kinetics of procaspase-8 cleavage. This probably does not allow the detection of products like CAP3 that are characterized by fast cleavage (Figure 3).
CAP3 is processed to p26 at the DISC
To find out whether CAP3 can be processed to p26 at the DISC we developed a Reverse in vitro caspase-8 cleavage assay. In this assay [35S]-labeled variants of the prodomains (aa 1–210, 1–216, 1–223) were added to lysates of anti-CD95-stimulated SKW6.4 cells before immunoprecipitation of the DISC (Figure 6a). Both, CAP3 as well as the 1–223 construct were processed to p26 in this assay. Thus, these data show that indeed CAP3 can be converted to p26. However, in this experiment the cleavage can take place both at the DISC and in the cytosol.
CAP3 is processed to p26 at the DISC. (a) Reverse in vitro caspase-8 cleavage assay. [35S]-labeled prodomains aa 1–210 (p26), 1–216 (CAP3) and 1–223 were incubated at 4°C overnight with the lysates of SKW6.4 cells stimulated with anti-APO-1. Afterwards DISCs were immunoprecipitated as in Figure 2. Samples were separated on 15% Anderson gels and developed by autoradiography. (b) Forward in vitro caspase-8 cleavage assays were performed using the same [35S]-labeled construct as in a. The assays were performed at 4°C overnight in the presence (lanes 7–12) or absence (lanes 1–6) of 50 μM zVAD-fmk. The beads and supernatants were separated, loaded on 15% Anderson gels and visualized by autoradiography (upper panel) and immunoblotting (lower panels). The DISC components were identified using anti-CD95, anti-FADD, anti-caspase-8 monoclonal antibodies (C15 and C5) recognizing the large and small subunits of caspase-8 (lanes 2, 4, 6)
To address the question if cleavage was taking place at the DISC a ‘standard’ in vitro caspase-8 cleavage assay was used (Figure 6b). DISCs isolated from SKW6.4 cells were incubated with in vitro translated, [35S]-labeled C-terminal deletion mutants of procaspase-8a (1–210, 1–216, 1–223). The analysis of both supernatants and DISC-containing beads (Figure 6b, lanes 1, 3, 5, upper panel) shows that the prodomains are cleaved at the DISC to p26 (Figure 6b, upper panel). The addition of zVAD-fmk blocks the processing of [35S]-labeled prodomains to p26 (Figure 6b, lanes 7–12, upper panel). It is likely that processing to p26 is performed by the activity of the mature caspase-8, which is present at the DISC (Figure 4d). Thus, CAP3 is processed at the DISC to p26 by caspase-8 activity that is inhibited by zVAD-fmk.
Discussion
Caspase-8 is a critical component of the CD95 apoptotic pathway.13, 21, 22 Activation of caspase-8 at the DISC plays a significant role in the regulation of CD95-mediated apoptosis. In this study, we investigated CAP3 and revealed new insights into the mechanism of procaspase-8 processing at the DISC. There have been several hypotheses as to the identity and function of CAP3, the most prominent hypothesis states that CAP3 is a short splice variant of procaspase-8 like procaspase-10c.11, 15, 23 We tried to find RNA encoding the entire open reading frame of CAP3 by 5′-RACE- polymerase chain reaction (PCR) (data not shown). However, these experiments did not result in the identification of another isoform of procaspase-8. By contrast, we present data that demonstrate that CAP3 is an intermediate product of procaspase-8 processing. We detected CAP3 only upon CD95 stimulation and procaspase-8 cleavage.
The detailed analysis of CAP3 using procaspase-8 deletion mutants has identified CAP3 as the prolonged prodomain of procaspase-8a that results from the cleavage at Asp216. In vitro cleavage assays with procaspase-8 mutants demonstrate that CAP3 is generated from procaspase-8a by cleavage at Asp216 and then converted to p26 by cleavage at Asp210.
Two isoforms of procaspase-8, procaspase-8a and -8b, were shown to be present at the DISC.10 Their autocatalytical processing undergoes similar steps involving the cleavage of p55/p53 to p43/p41 that is followed by the processing of p43/p41 to p26/p24. Interestingly, we also observed a product, which very likely corresponds to CAP3b, regarding its molecular mass. However, the detection of CAP3b was often complicated, as the corresponding lane overlaps with the one from p26. Futhermore, in some cell lines, we observed a different ratio between CAP3b and CAP3a, including some cases when CAP3b was hardly detectable in contrast to CAP3a. This indicates that procaspases-8a and -8b might have different kinetics of processing. This question should be addressed in future studies.
CAP3 processing takes place very quickly after CD95 stimulation. This was shown both by a kinetic analysis of CAP3 presence at the DISC and by an in vitro caspase-8 cleavage assay. In this assay CAP3 was detectable only when the procaspase-8a mutant D210A was used, the generation of p26 was not possible and the cleavage was stopped at Asp216. The fact that CAP3 could not be detected by an in vitro cleavage assay for wild-type procaspase-8a has always been a serious argument against CAP3 being a proteolytic product of procaspase-8. It seems likely, that the kinetics of in vitro cleavage assay is different from that in vivo. Therefore, experimental conditions may not allow the detection of products with a fast conversion rate. Thus, our data show that CAP3 is an intermediate product of procaspase-8 processing that is quickly processed at the DISC.
The analysis of procaspase-8 processing at the DISC in the presence of covalent inhibitors zVAD-fmk as well as zIETD-fmk confirmed that CAP3 is a product of procaspase-8 processing. The amount of CAP3 at the DISC was enhanced upon addition of the inhibitors. The generation of CAP3 occurs possibly due to the incomplete inhibition of caspase-8 activity by zVAD-fmk. This might be due to spatial nonaccessibility of active sites of procaspase-8 at the DISC.
Another question to address with respect to the mechanism of CAP3 formation was if there is another protease that might generate CAP3 at the DISC. Procaspase-10 is the only other procaspase that is reported to be present at the DISC so far.14 However, the generation of CAP3 by procaspase-10 is rather unlikely as CAP3 is present at the DISC of Raji cells, which are lacking procaspase-10 (Figure 1). In addition, we examined the influence of cathepsin or granzyme inhibitors on CAP3 formation but did not see any effect (data not shown). Thus, we believe that there is no other protease responsible for the generation of CAP3.
Based on our experimental data, we propose the following model of procaspase-8 activation at the DISC. First, procaspase-8 is recruited to the DISC via homophilic interactions with the DED of FADD (Figure 7). There is strong evidence from a number of in vitro studies that dimer formation of two procaspase-8 molecules results in their activation followed by processing.18, 24, 25, 26 The active procaspase-8 dimer may cross-cleave another procaspase-8 dimer. Cleavage steps for procaspase-8a involve the residues Asp374, Asp384 and Asp216. As a result of the cleavage steps CAP3, p18 and the p10 subunits are formed. The formed heterotetramer of mature caspase-8 quickly accomplishes procaspase-8 processing, for example, converting CAP3 to p26 by cleavage at Asp210.
Model of procaspase-8 activation. Upon oligomerization at the DISC, procaspase-8 molecules form dimers. Dimerization of two procaspase-8 molecules results in their activation. The activated procaspase-8 dimer performs cross-cleavage of another dimer. For procaspase-8a processing occurs at Asp374, Asp384 and Asp216 (The positions of Asp (D) residues are indicated). As the result of those cleavage steps p43, CAP3, p18 and p10 cleavage products are generated. The activity of mature caspase-8 processes CAP3 to p26. This step is inhibited by zVAD-fmk
According to previous publications and our analysis of procaspase-8 processing at the DISC (Figure 3a and b), the DED-containing prodomains p24 and p26 remain bound to the DISC for hours after stimulation.10 The remaining DED – containing prodomains at the DISC are very likely playing an important role. It was demonstrated in overexpression studies that prodomains of procaspase-8 can induce apoptosis without stimulation of CD95 (APO-1/Fas).27 When fused to green fluorescent protein (GFP) each DED of procaspase-8, and also FADD-DED, were shown to form death effector filaments. FADD and procaspase-8 were recruited to these death effector filaments and activated. Since prodomains remain bound to the DISC they might form death-effector-filament-like structures that could serve as a ‘platform’ for the recruitment of many procaspase-8 molecules enhancing the formation of mature caspase-8.
To get more functional insight into the importance of CAP3 in the DISC and the processing of procaspase-8, we tried to reconstitute caspase-8 deficient Jurkat A3 cells as well as 293T cells with caspase-8 at a reduced level.28 Cells were transfected with wild-type procaspase-8a as well as with procaspase-8a mutants (D210A, D216A, D210A/D216A). However, we failed to get any processing of the wild type procaspase-8a after reconstitution, neither in transient nor in stable transfectants pointing towards a defect in these cell lines interfering with processing of procaspase-8.
To understand the functional role of CAP3 and to compare it to p26, we generated stable BJAB cell lines overexpressing both, prodomains aa 1–216 and aa 1–210 (data not shown). However, we did not observe any differences between those cell lines with respect to the sensitivity towards CD95-induced apoptosis. DISC formation was observed at the same rate. Considering the structural similarity of these two prodomains, a redundant function in CD95 signaling was expected.
In this study, we examined procaspase-8 processing directly at the DISC, in contrast to many other studies where procaspase-8 cleavage steps were monitored using artificial drug-induced dimerization of procaspase-8. We revealed an additional step in procaspase-8 processing, that is, the generation of CAP3 as a prolonged prodomain of procaspase-8 that is quickly converted to p26. It is essential to understand all processing steps of procaspase-8 at the DISC to eventually regulate CD95-induced apoptosis. In addition, these data might have further implications for the development of specific CD95 apoptosis inhibitors. Thus, these findings might be a basis for drug design in diseases connected with the dysregulation in CD95 signaling such as multiple sclerosis, Alzheimer's disease, diabetes mellitus, rheumatoid arthritis, Hodgkin's disease and different cancers.
Experimental Procedures
Cell lines
The T-cell lines HUT78, CEM, Jurkat (Caspase-8 deficient, clone I9.2), Jurkat clone A3, Jurkat clone J16, H9 and the B lymphoblastoid cell line SKW6.4 were maintained in RPMI 1640 (Life Technologies, Germany), 10 mM HEPES (Life Technologies, Germany), 50 μg/ml Gentamycin (Life Technologies, Germany), 10% fetal calf serum (Life Technologies, Germany) in 5% CO2. The caspase-8 deficient Jurkat cell line (clone I9.2) and Jurkat clone A3 were obtained from J Blenis (Harvard Medical School).
Antibodies and reagents
Anti-FADD mAb (mouse IgG1) was purchased from Transduction Laboratories (Lexington, KY, USA). The anticaspase-8 mAb C15 (mouse IgG2b) recognizes the p18 subunit of caspase-8. The C5 mAb (mouse IgG2a) recognizes the p10 subunit of caspase-8, and the anticaspase-8 monoclonal antibodies N1 and N2 (both mouse IgG1) recognize the prodomain of procaspase-8.10 Anti-APO-1 is an agonistic mAb (IgG3) recognizing an epitope on the extracellular part of CD95 (APO-1/Fas).7 The horseradish peroxidase-conjugated goat anti-mouse IgG1, −2a and −2b were from Southern Biotechnology Associates (United Kingdom). Leucine zipper (LZ)-CD95L was produced as described.29 Cell permeable pan-caspase inhibitors zVAD-fmk and Biotin-VAD-fmk were purchased from Merck (Germany). All other chemicals used were of analytical grade and purchased from Merck (Germany) or Sigma (Germany).
Cloning of procaspase-8 mutants and prodomains
The prodomains were cloned into the pEF4 expression vector (Invitrogen, Germany) using the polymerase chain reaction and the following primers: Procaspase-8 sense: 5′-ggggtacccc-ATGGACTTCAGCAGAAATC-3′, 1–223 procaspase-8 antisense: 5′-gctctagagcctaGTCCA-AAGTCTGTGATTC-3′, 1–216 procaspase-8 antisense: 5′-gctctagagcctaATCCTGTTCTCTTG-GAGAG-3′, 1–210 procaspase-8 antisense: 5′-gctctagagcctaGTCCGAGATTGTCATTACC-3′. Procaspase-8 mutants were cloned via overlap-PCR into the pEF4 expression vector (Invitrogen, Germany). The N-terminal part of procaspase-8 was amplified using the following Forward-Primer: 5′-ggggtaccccATGGACTTCAGCAGAA-ATC-3′ and as Reverse-Primers: D210A: 5′-CTTGGAGAGGCCGAGATTG-3′, D216A: 5′-GATTCACTAGCCTGTTCTC-3′, D374A: 5′-CCTCTGAAGCAGTCTCAAC-3′, D384A: 5′-GGTGATGATAAAGCCATTTC-3′. The C-terminal part of procaspase-8 was amplified using the following Reverse-Primer: 5′-gctctagagcTTAATCAGAAGGGAAAAGT-3′ and as Forward-Primers: D210A: 5′-CAATCTCGGCCTCTCCAAG-3′, D216A: 5′-GAGAACAGGCTAGTGAATC-3′, D374A: 5′-GTTGAGACTGCTTCAGAGG-3′, D384A: 5′-GAAATGGCTTTATCATCACC-3′.
DISC analysis by immunoprecipitation
The composition of the CD95 DISC was determined as follows. Cells (1 × 108) were either treated with 10 μg of LZ-CD95L-containing supernatant or 2 μg/ml anti-APO-1 (IgG3) for 5 min (unless otherwise indicated) at 37°C, washed twice in 1 × phosphate-buffered saline (PBS) and subsequently lysed in lysis buffer (30 mM Tris/HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (Sigma, Germany), protease inhibitor cocktail (Roche, Switzerland), 1% Triton X-100 (Serva, Germany), and 10% glycerol) (stimulated condition) or lysed without treatment (unstimulated condition). The CD95 DISC was immunoprecipitated at 4°C for 2 h or overnight with 2 μg of anti-APO-1 coupled to Protein-A Sepharose (when cells were stimulated with LZ-CD95L) or with Protein-A Sepharose beads (when cells were stimulated with anti-APO-1). For N1-immunoprecipitation anticaspase-8 mAb N1 was covalently coupled to CNBr-activated Sepharose CL-4B (Pharmacia, Sweden). N1-Sepharose beads (30 μl) were used for immunoprecipitation at 4°C for 2 h. All beads were washed five times with 20 volumes of lysis buffer. The immunoprecipitates were analyzed either using 2-D isoelectric focusing (IEF)/SDS-PAGE as described30 or 1-D gels. The gels were transferred to Hybond nitrocellulose membrane (Amersham Pharmacia Biotech, Germany), blocked with 5% nonfat dry milk in PBS/Tween (PBS plus 0.05% Tween 20) for 1 h, washed with PBS/Tween, and incubated with the primary antibody in PBS/Tween at 4°C overnight. Blots were developed with a chemoluminescence method following the manufacturer's protocol (Perkin Elmer Life Sciences, Germany).
In vitro translation and Forward- and Reverse in vitro caspase-8 cleavage assays
The CD95 DISC was immunoprecipitated from 5 × 107 SKW6.4 cells and immunoprecipitates (Forward in vitro caspase-8 cleavage assay) were incubated with in vitro translated [35S]-labeled procaspase-8 prodomains or mutants (TNT, T7 coupled reticulocyte lysate system, Promega) in caspase-8 cleavage buffer (50 mM HEPES, pH 7.4, 100 mM NaCl, 0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid (CHAPS), 10 mM dithiothreitol and 10% sucrose) at 4°C overnight (unless otherwise indicated in the text).11 For Reverse in vitro caspase-8 cleavage assay in vitro translated [35S]-labeled procaspase-8 prodomains were incubated with stimulated lysates at 4°C overnight and CD95 DISC immunoprecipitation was done afterwards. Cleavage reactions were separated on 15% Anderson gels, blotted and subjected to autoradiography.
Abbreviations
- DISC:
-
death-inducing signaling complex
- DED:
-
death effector domain
- FADD:
-
Fas-associated DED
- DD:
-
death domain
- c-FLIP:
-
c-FLICE inhibitory protein
- mAb:
-
monoclonal antibody
- PBS:
-
phosphate-buffered saline
- CHAPS:
-
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- CAP:
-
cytotoxicity-associated protein
- aa:
-
amino acids
- ESI-MS/MS:
-
electrospray ionization-tandem mass spectrometry
- pI:
-
isoelectric point
- min:
-
minutes
- h:
-
hours
- PCR:
-
polymerase chain reaction
- GFP:
-
green fluorescent protein
- IEF:
-
isoelectric focussing
- LZ-CD95L:
-
leucine zipper-CD95 ligand
- 1-D:
-
one-dimensional
- 2-D:
-
two-dimensional
- RT:
-
room temperature
- Asp:
-
aspartate
- zVAD-fmk:
-
benzoyl-valine-alanine-aspartate-fluoromethylketone
- zIETD-fmk:
-
benzoyl-isoleucine-glutamate-threonine-aspartate-fluoromethylketone
References
Krammer PH (2000) CD95's deadly mission in the immune system. Nature 407: 789–795
Walczak H and Krammer PH (2000) The CD95 (APO-1/Fas) and the TRAIL (APO-2L) apoptosis systems. Exp. Cell. Res. 256: 58–66
Schmitz I, Kirchhoff S and Krammer PH (2000) Regulation of death receptor-mediated apoptosis pathways. Int. J. Biochem. Cell Biol. 32: 1123–1136
Nagata S (1997) Apoptosis by death factor. Cell 88: 355–365
Peter ME and Krammer PH (2003) The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 10: 26–35
Suda T, Takahashi T, Golstein P and Nagata S (1993) Molecular cloning and expression of the Fas ligand, a novel member of the tumor necrosis factor family. Cell 75: 1169–1178
Trauth BC, Klas C, Peters AM, Matzku S, Moller P, Falk W, Debatin KM and Krammer PH (1989) Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science 245: 301–305
Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH and Peter ME (1995) Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 14: 5579–5588
Golks A, Brenner D, Fritsch C, Krammer PH and Lavrik IN (2005) c-FLIPR, a new regulator of death receptor-induced apoptosis. J. Biol. Chem. 280: 14507–14513
Scaffidi C, Medema JP, Krammer PH and Peter ME (1997) FLICE is predominantly expressed as two functionally active isoforms, caspase-8/a and caspase-8/b. J. Biol. Chem. 272: 26953–26958
Medema JP, Scaffidi C, Kischkel FC, Shevchenko A, Mann M, Krammer PH and Peter ME (1997) FLICE is activated by association with the CD95 death-inducing signaling complex (DISC). EMBO J. 16: 2794–2804
Lavrik I, Krueger A, Schmitz I, Baumann S, Weyd H, Krammer PH and Kirchhoff S (2003) The active caspase-8 heterotetramer is formed at the CD95 DISC. Cell Death Differ. 10: 144–145
Muzio M, Chinnaiyan AM, Kischkel FC, O'Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME and Dixit VM (1996) FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death--inducing signaling complex. Cell 85: 817–827
Sprick M, Rieser E, Stahl H, Grosse-Wilde A, Weigand M and Walczak H (2002) Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J. 21: 4520–4530
Yang X, Chang HY and Baltimore D (1998) Autoproteolytic activation of pro-caspases by oligomerization. Mol. Cell. 1: 319–325
Anderson NL and Anderson NG (1978) Analytical technique for cell fractions. XXII. Two-dimensional analysis of serum and tissue proteins. Multiple gradient-slab gel electrophoresis. Anal. Biochem. 85: 341–354
Sakamaki K, Tsukumo S and Yonehara S (1998) Molecular cloning and characterization of mouse caspase-8. Eur. J. Biochem. 253: 399–405
Chang DW, Xing Z, Capacio VL, Peter ME and Yang X (2003) Interdimer processing mechanism of procaspase-8 activation. EMBO J. 22: 4132–4142
Chang DW and Yang X (2003) Activation of procaspases by FK506 binding protein-mediated oligomerization. Sci. STKE. 2003: PL1
Kischkel FC, Lawrence DA, Tinel A, LeBlanc H, Virmani A, Schow P, Gazdar A, Blenis J, Arnott D and Ashkenazi A (2001) Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J. Biol. Chem. 2: 2
Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL, Rebrikov D, Brodianski VM, Kemper OC, Kollet O, Lapidot T, Soffer D, Sobe T, Avraham KB, Goncharov T, Holtmann H, Lonai P and Wallach D (1998) Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 9: 267–276
Juo P, Kuo CJ, Yuan J and Blenis J (1998) Essential requirement for caspase-8/FLICE in the initiation of the Fas-induced apoptotic cascade. Curr. Biol. 8: 1001–1008
Sprick MR, Rieser E, Stahl H, Grosse-Wilde A, Weigand MA and Walczak H (2002) Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J. 21: 4520–4530
Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR and Salvesen GS (2003) A unified model for apical caspase activation. Mol. Cell. 11: 529–541
Muzio M, Stockwell BR, Stennicke HR, Salvesen GS and Dixit VM (1998) An induced proximity model for caspase-8 activation. J. Biol. Chem. 273: 2926–2930
Martin DA, Siegel RM, Zheng L and Lenardo MJ (1998) Membrane oligomerization and cleavage activates the caspase-8 (FLICE/MACHalpha1) death signal. J. Biol. Chem. 273: 4345–4349
Siegel RM, Martin DA, Zheng L, Ng SY, Bertin J, Cohen J and Lenardo MJ (1998) Death-effector filaments: novel cytoplasmic structures that recruit caspases and trigger apoptosis. J. Cell. Biol. 141: 1243–1253
Kataoka T and Tschopp J (2004) N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol. Cell. Biol. 24: 2627–2636
Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C, Smolak P, Goodwin RG, Rauch CT, Schuh JC and Lynch DH (1999) Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 5: 157–163
Huber LA, Ullrich O, Takai Y, Lutcke A, Dupree P, Olkkonen V, Virta H, de Hoop MJ, Alexandrov K and Peter M (1994) Mapping of Ras-related GTP-binding proteins by GTP overlay following two-dimensional gel electrophoresis. Proc. Natl. Acad. Sci. USA 91: 7874–7878
Acknowledgements
We thank Drs Sabine Kirchhoff and Frank Kischkel for the contribution to the project, Christine Stumpf and Wolfgang Müller for technical assistance, Drs Karsten Gülow, Elisabeth Suri-Payer, Rüdiger Arnold and Dagmar Riess for discussion and Heidi Sauter for expert secretarial assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Deutsche Krebshilfe, the European Community and the Wilhelm Sander Stiftung.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by G Salvesen
Rights and permissions
About this article
Cite this article
Golks, A., Brenner, D., Schmitz, I. et al. The role of CAP3 in CD95 signaling: new insights into the mechanism of procaspase-8 activation. Cell Death Differ 13, 489–498 (2006). https://doi.org/10.1038/sj.cdd.4401766
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401766
Keywords
This article is cited by
-
Dissecting DISC regulation via pharmacological targeting of caspase-8/c-FLIPL heterodimer
Cell Death & Differentiation (2020)
-
Long and short isoforms of c-FLIP act as control checkpoints of DED filament assembly
Oncogene (2020)
-
Modulation of CD95-mediated signaling by post-translational modifications: towards understanding CD95 signaling networks
Apoptosis (2019)
-
Molecular architecture of the DED chains at the DISC: regulation of procaspase-8 activation by short DED proteins c-FLIP and procaspase-8 prodomain
Cell Death & Differentiation (2016)
-
Systems biology of death receptor networks: live and let die
Cell Death & Disease (2014)