Abstract
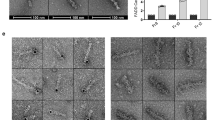
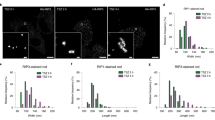
The assembly of the death-inducing signaling complex (DISC) and death effector domain (DED) filaments at CD95/Fas initiates extrinsic apoptosis. Procaspase-8 activation at the DED filaments is controlled by short and long c-FLIP isoforms. Despite apparent progress in understanding the assembly of CD95-activated platforms and DED filaments, the detailed molecular mechanism of c-FLIP action remains elusive. Here, we further addressed the mechanisms of c-FLIP action at the DISC using biochemical assays, quantitative mass spectrometry, and structural modeling. Our data strongly indicate that c-FLIP can bind to both FADD and procaspase-8 at the DED filament. Moreover, the constructed in silico model shows that c-FLIP proteins can lead to the formation of the DISCs comprising short DED filaments as well as serve as bridging motifs for building a cooperative DISC network, in which adjacent CD95 DISCs are connected by DED filaments. This network is based on selective interactions of FADD with both c-FLIP and procaspase-8. Hence, c-FLIP proteins at the DISC control initiation, elongation, and composition of DED filaments, playing the role of control checkpoints. These findings provide new insights into DISC and DED filament regulation and open innovative possibilities for targeting the extrinsic apoptosis pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Krammer PH, Arnold R, Lavrik IN. Life and death in peripheral T cells. Nat Rev Immunol. 2007;7:532–42.
Lavrik IN, Krammer PH. Regulation of CD95/Fas signaling at the DISC. Cell Death Differ. 2012;19:36–41.
Dickens LS, Boyd RS, Jukes-Jones R, Hughes MA, Robinson GL, Fairall L, et al. A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Mol Cell. 2012;47:291–305.
Fu TM, Li Y, Lu A, Li Z, Vajjhala PR, Cruz AC, et al. Cryo-EM structure of caspase-8 tandem DED filament reveals assembly and regulation mechanisms of the death-inducing signaling complex. Mol Cell. 2016;64:236–50.
Schleich K, Warnken U, Fricker N, Ozturk S, Richter P, Kammerer K, et al. Stoichiometry of the CD95 death-inducing signaling complex: experimental and modeling evidence for a death effector domain chain model. Mol Cell. 2012;47:306–19.
Ozturk S, Schleich K, Lavrik IN. Cellular FLICE-like inhibitory proteins (c-FLIPs): fine-tuners of life and death decisions. Exp Cell Res. 2012;318:1324–31.
Golks A, Brenner D, Fritsch C, Krammer PH, Lavrik IN. c-FLIPR, a new regulator of death receptor-induced apoptosis. J Biol Chem. 2005;280:14507–13.
Chang DW, Xing Z, Pan Y, Algeciras-Schimnich A, Barnhart BC, Yaish-Ohad S, et al. c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 2002;21:3704–14.
Micheau O, Thome M, Schneider P, Holler N, Tschopp J, Nicholson DW, et al. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J Biol Chem. 2002;277:45162–71.
Peter ME. The flip side of FLIP. Biochemical J. 2004;382:e1–3.
Thome M, Tschopp J. Regulation of lymphocyte proliferation and death by FLIP. Nat Rev Immunol. 2001;1:50–58.
Yu JW, Jeffrey PD, Shi Y. Mechanism of procaspase-8 activation by c-FLIPL. Proc Natl Acad Sci USA. 2009;106:8169–74.
Fricker N, Beaudouin J, Richter P, Eils R, Krammer PH, Lavrik IN. Model-based dissection of CD95 signaling dynamics reveals both a pro- and antiapoptotic role of c-FLIPL. J Cell Biol. 2010;190:377–89.
Hughes MA, Powley IR, Jukes-Jones R, Horn S, Feoktistova M, Fairall L, et al. Co-operative and hierarchical binding of c-FLIP and caspase-8: a unified model defines how c-flip isoforms differentially control cell fate. Mol Cell. 2016;61:834–49.
Schleich K, Buchbinder JH, Pietkiewicz S, Kahne T, Warnken U, Ozturk S, et al. Molecular architecture of the DED chains at the DISC: regulation of procaspase-8 activation by short DED proteins c-FLIP and procaspase-8 prodomain. Cell Death Differ. 2016;23:681–94.
Scaffidi C, Schmitz I, Krammer PH, Peter ME. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem. 1999;274:1541–8.
Hwang EY, Jeong MS, Park SY, Jang SB. Evidence of complex formation between FADD and c-FLIP death effector domains for the death inducing signaling complex. BMB Rep. 2014;47:488–93.
Ueffing N, Keil E, Freund C, Kuhne R, Schulze-Osthoff K, Schmitz I. Mutational analyses of c-FLIPR, the only murine short FLIP isoform, reveal requirements for DISC recruitment. Cell Death Differ. 2008;15:773–82.
Majkut J, Sgobba M, Holohan C, Crawford N, Logan AE, Kerr E, et al. Differential affinity of FLIP and procaspase 8 for FADD’s DED binding surfaces regulates DISC assembly. Nat Commun. 2014;5:3350.
Golks A, Brenner D, Schmitz I, Watzl C, Krueger A, Krammer PH, et al. The role of CAP3 in CD95 signaling: new insights into the mechanism of procaspase-8 activation. Cell Death Differ. 2006;13:489–98.
Hoffmann JC, Pappa A, Krammer PH, Lavrik IN. A new C-terminal cleavage product of procaspase-8, p30, defines an alternative pathway of procaspase-8 activation. Mol Cell Biol. 2009;29:4431–40.
Juo P, Kuo CJ, Yuan J, Blenis J. Essential requirement for caspase-8/FLICE in the initiation of the Fas-induced apoptotic cascade. Curr Biol. 1998;8:1001–8.
Sprick MR, Rieser E, Stahl H, Grosse-Wilde A, Weigand MA, Walczak H. Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J. 2002;21:4520–30.
Lavrik IN. Systems biology of death receptor networks: live and let die. Cell Death Dis. 2014;5:e1259.
Bentele M, Lavrik I, Ulrich M, Stosser S, Heermann DW, Kalthoff H, et al. Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J Cell Biol. 2004;166:839–51.
Warnken U, Schleich K, Schnolzer M, Lavrik I. Quantification of high-molecular weight protein platforms by AQUA mass spectrometry as exemplified for the CD95 death-inducing signaling complex (DISC). Cells. 2013;2:476–95.
Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol Cell Proteom. 2012;11:202–14.
Chaudhury S, Berrondo M, Weitzner BD, Muthu P, Bergman H, Gray JJ. Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS ONE. 2011;6:e22477.
Lavrik I, Golks A, Krammer PH. Death receptor signaling. J Cell Sci. 2005;118:265–7.
Algeciras-Schimnich A, Shen L, Barnhart BC, Murmann AE, Burkhardt JK, Peter ME. Molecular ordering of the initial signaling events of CD95. Mol Cell Biol. 2002;22:207–20.
Algeciras-Schimnich A, Peter ME. Actin dependent CD95 internalization is specific for Type I cells. FEBS Lett. 2003;546:185–8.
Horn S, Hughes MA, Schilling R, Sticht C, Tenev T, Ploesser M, et al. Caspase-10 negatively regulates caspase-8-mediated cell death, switching the response to CD95L in favor of NF-kappaB activation and cell survival. Cell Rep. 2017;19:785–97.
Neumann L, Pforr C, Beaudouin J, Pappa A, Fricker N, Krammer PH, et al. Dynamics within the CD95 death-inducing signaling complex decide life and death of cells. Mol Syst Biol. 2010;6:352.
Schmidt JH, Pietkiewicz S, Naumann M, Lavrik IN. Quantification of CD95-induced apoptosis and NF-kappaB activation at the single cell level. J Immunol Methods. 2015;423:12–17.
Pietkiewicz S, Eils R, Krammer PH, Giese N, Lavrik IN. Combinatorial treatment of CD95L and gemcitabine in pancreatic cancer cells induces apoptotic and RIP1-mediated necroptotic cell death network. Exp Cell Res. 2015;339:1–9.
Pietkiewicz S, Schmidt JH, Lavrik IN. Quantification of apoptosis and necroptosis at the single cell level by a combination of Imaging Flow Cytometry with classical Annexin V/propidium iodide staining. J Immunol Methods. 2015;423:99–103.
MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–8.
Acknowledgements
We acknowledge Volkswagen Foundation (VW 90315), Wilhelm Sander-Stiftung (2017.008.01), Center of dynamic systems (CDS), funded by the EU-program ERDF (European Regional Development Fund) and DFG (LA 2386) for supporting our work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Hillert, L.K., Ivanisenko, N.V., Espe, J. et al. Long and short isoforms of c-FLIP act as control checkpoints of DED filament assembly. Oncogene 39, 1756–1772 (2020). https://doi.org/10.1038/s41388-019-1100-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-1100-3
This article is cited by
-
MIND bomb 2 prevents RIPK1 kinase activity-dependent and -independent apoptosis through ubiquitylation of cFLIPL
Communications Biology (2021)
-
Cryo-EM structural analysis of FADD:Caspase-8 complexes defines the catalytic dimer architecture for co-ordinated control of cell fate
Nature Communications (2021)
-
Pharmacological targeting of c-FLIPL and Bcl-2 family members promotes apoptosis in CD95L-resistant cells
Scientific Reports (2020)