Abstract
Splenic marginal zone lymphoma (SMZL) is a rare non-Hodgkin’s lymphoma that recently has been recognized as an entity. The first goal of this study was to identify potential chromosomal aberrations in this entity by cytogenetic analysis and comparative genomic hybridization (CGH). The second goal was to assess the frequency of 7q31–32 allelic imbalances in SMZL with primary involvement of the spleen and the typical immunophenotype (IgM+; IgDdim; and CD5−, CD10−, and CD23−). We applied CGH and cytogenetics to 13 cases of SMZL with primary splenic involvement. By CGH, we found DNA copy number changes in 11 of 13 cases. Overall chromosomal gains were more frequent than chromosomal losses. Gains were most frequently detected for chromosome X, chromosome 3, and chromosome 18. Losses commonly involved chromosome 7 and chromosome 6.CGH and cytogenetic analysis showed a deletion in chromosome 7q31 in 4 cases. Loss of heterozygosity (LOH) analysis using three microsatellite markers located at 7q31 revealed LOH in 9 cases. Remarkably, 2 of the 4 cases that lacked a 7q31 deletion had an atypical immunophenotype because they were partially CD23 positive. The other 2 cases were not informative. The findings indicate that SMZL with primary splenic presentation and the typical IgM+, IgDdim, CD5−, CD10−, CD23− immunophenotype is characterized by the presence of deletions in chromosome 7q31–32.
Similar content being viewed by others
Main
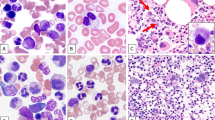
Splenic marginal zone lymphoma (SMZL) is a rare low-grade B-cell neoplasm that occurs mostly in middle-aged and elderly patients (1). Similar cases had been described before as lymphocytic lymphoma with predominant splenomegaly (2, 3). In the WHO classification, SMZL has been recognized as an entity with immunophenotypical features that are different from those of the other low-grade B-cell lymphomas (WHO). Most cases are IgM+ and IgDdim and lack expression of CD5, CD23, and CD10 (1). The histological features of SMZL include infiltration of small atypical lymphocytes in the mantle zone and of medium-sized lymphocytes with pale cytoplasm and oval clear nucleus in the marginal zone, resulting in a mixed mantle-zone and marginal-zone involvement pattern. Some cases exhibit follicular colonization. Overall, there is a clear increase of white pulp, with more follicles per square area than normally seen, and red pulp infiltration also is frequently seen. Patients with SMZL frequently have bone marrow and peripheral blood involvement (4).
Analysis of SMZL by cytogenetic and FISH approaches has revealed gains of chromosome 3 and 18; losses of chromosome 1, 3p, and 7q; and structural abnormalities of chromosome 8 and 14 (5, 6, 7). CGH (comparative genomic hybridization) analysis of SMZL confirmed these results and in addition revealed gains of chromosome 5, 12, and 20 and losses of chromosome 17p (8, 9).
We performed cytogenetic and CGH analyses and a loss of heterozygosity (LOH) assay for the 7q31–32 region of 13 cases of SMZL with primary or exclusive involvement of spleen.
MATERIALS AND METHODS
Patient Samples
Thirteen cases of SMZL presenting with splenomegaly were selected for this study. These cases were diagnosed at the University Hospital Groningen, Groningen, The Netherlands or at the Cross Cancer Institute, Edmonton, Canada between 1990 and 2000. Cases with CD5 positivity or strong CD23 positivity were excluded. Two cases with only partial, weak CD23 positivity were included.
Immunohistochemistry was performed on frozen sections using standard protocols and the monoclonal antibodies anti-CD20, anti-CD5, anti-CD10, anti-IgM, anti-IgD, anti-κ (all from Dakopatts, Glostrup, Denmark), anti-λ (Becton-Dickinson, Franklin Lakes, NJ), and anti-CD23 (Novocastra Laboratories Ltd., Newcastle upon Tyne, United Kingdom). Relevant clinical data were retrieved from the medical records (see Table 1). Peripheral blood smears of all patients were analyzed for atypical lymphocytes. In 11 cases, atypical and/or clonal cells were detected in the peripheral blood using flow cytometry, morphology, or both (see Table 2). In two cases, no tumor cells were detected in the peripheral blood.
Cytogenetic Studies
A piece of macroscopically involved tissue was washed through with RPMI-1640 (supplemented with 15% FCS, glutamine, and antibiotics) and subsequently cut into small pieces in a small Petri dish filled with RPMI. The suspension was transferred to a tube and centrifuged for 10 minutes at 2000 × g. About 25 × 106 cells (counted on a cell counter) were added to 10 mL of culture medium (at least 2 cultures per case) and incubated for 24 hours in a 5% CO2 incubator at 37° C. Cells were harvested after 24 hours, and chromosome preparations were made using standard cytogenetic techniques. The chromosomes were G-banded using Pancreatin 0.1% and Giemsa 10%. The remaining cell suspension was stored at −20° C. Cytogenetic analysis and the karyotype description was performed according to the ISCN 1995 (ISCN [1995]: An international system for human cytogenetic nomenclature, F.Mitelman, ed. S.Karger, Basel).
Metaphase Preparations for CGH
Normal metaphase spreads for CGH were prepared from phytohemagglutinin-stimulated cultured blood from healthy male and female individuals by standard procedures of colcemid arrest, hypotonic treatment, and methanol–acetic acid (3:1, v/w) fixation. Metaphase spreads were prepared fresh 1 day before use.
CGH
DNA was extracted from frozen tumor tissue blocks and from the full blood of healthy individuals using salt–chloroform extraction after proteinase K–SDS digestion according to standard methods. All tissue blocks used contained >70% tumor cells.
CGH was performed as described by Kallioniemi and co-workers (10) with some adjustments. Approximately 1 μg of tumor DNA (test DNA) and 1 μg of normal DNA (reference DNA) were labeled by nick translation using a biotin and digoxigenin labeling kit; Roche Molecular Biochemicals, Mannheim, Germany) with Biotin-16-dUTP and digoxigenin-11-dUTP. The size of the labeled fragments ranged from 400–3000 bp, as assessed by agarose gel electrophoreses. Aliquots of 400 ng of labeled test DNA and control DNA were ethanol precipitated with 50-μg unlabeled human Cot1 DNA (Invitrogen BV, Breda, The Netherlands) and 10 μg of salmon sperm DNA (Sigma). The DNA mixture was dissolved in 15 μL of hybridization mixture (50% deionized formamide; 2 × SSC; 10% dextran sulfate, pH 7.0). The hybridization mixture with the labeled DNA was applied to a normal male or female metaphase slide. A cover clip was added, and the slide was sealed with rubber cement. The slide with the DNA hybridization mixture was denatured at 74° C for 3 minutes and hybridized at 37° C during 72 hours. Posthybridization washes (three 5-min washes in 4 × SSC at 42° C and three 5-min washes in 0.1 SSC at 60° C) were performed before immunological detection. For immunological detection the slide was incubated with 10 μL of streptavidin-FITC (Roche Molecular Biochemicals, Mannheim, Germany) and 10 μL of anti–DIG-TRITC (Roche Molecular Biochemicals) in 4 × SSC, Tween-20, and 1% fat-free powdered milk over a 1-hour period to detect the biotin-labeled tumor DNA and digoxigenin-labeled normal DNA. After washing and dehydration, the slides were mounted with antifade solution containing 4,6-diamidino-2-phenylindole (DAPI) as counterstain (Vectashield; Vector laboratories, Burlingame, CA).
The grayscale images of the three different fluorochromes were captured using a Leica DMRA fluorescence microscope that was equipped with DAPI, FITC, and TRITC filters (Chroma, Brattleboro, VT); CCD camera (Cohu 4912 CCD camera, San Diego, CA); and the image-capturing program QFISH (Leica, Cambridge, England). The three captured images were combined, and pseudocolor was applied matching the original colors of the fluorochromes. The reversed DAPI image was used for chromosome identification. The ratio between the FITC (tumor) and TRITC (normal) fluorescence was calculated with use of the QCGH software program (Leica). For each case the mean of the individual ratio profiles of 6–10 metaphase spreads was calculated. The three vertical lines at the right side of the ideogram (Fig. 1) represent the balanced state of the chromosomal copy number. The middle line was set at ratio 1.0, the upper threshold (right) was set at 1.15 for gain, and the lower threshold (left) was set at 0.85 for loss, as determined by control experiments using two normal DNA samples. Overrepresentations were considered as high-level amplifications when the fluorescence ratio values were >2.0. The centromeric regions; heterochromatin blocks of chromosomes 1, 9, and 16; the satellite regions of the acrocentric chromosomes; and the Y chromosome were excluded from analysis because of the abundance of highly repetitive DNA sequences.
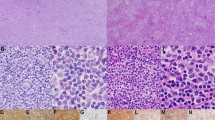
CGH ratio profile on chromosome 7 of Cases 2, 3, 6, and 81 with deletion of 7q31. Blue line represents ratio between normal control DNA, labeled red, with tumor DNA, labeled green. Ratio less than red line, which is 0.85 = loss of tumor DNA; ratio more than green line, which is 1.15 = gain of tumor DNA.
LOH Analysis
Three microsatellite markers on chromosome 7q31–32 were used: D7S686, D7S685, and D7S487. The three microsatellite markers were selected based on their location (Genethon map release) (11) and on the frequent LOH detected in SMZL (12). For each primer set, one of the primers was labeled with either 5′-6-FAM (Biolegio, Malder, The Netherlands) or 5′-HEX (Biolegio). PCR reactions were performed in a final volume of 20 μL containing approximately 50 ng of template DNA, 4 mmol dNTPs, 2.5 mmol/L Mg2CL, 75 pmol of each primer, and 1 U of Taq polymerase (Amersham Pharmacia Biotech, Uppsala, Sweden). PCR was performed on a PTC-225 thermal cycler (MJ research, Waltman, MA). Amplification consisted of an initial denaturing of 4 minutes at 95° C, 30 cycles of 30 sec at 95° C, 30 sec at 57° C, and 30 sec at 72° C, followed by an additional step of 10 minutes at 72° C. We mixed 2.3 μL of the PCR products with 2.5 μL of deionized formamide and 0.2 μL of ET-400R size standard (Amersham Pharmacia Biotech) and separated on a MegaBACE 1000 capillary sequencer (Amersham Pharmacia Biotech). Results were analyzed using the Genetic Profiler v1.1 (Amersham Pharmacia Biotech). Allelic imbalance (AI) was determined by comparing the peak ratios of both alleles of the tumor DNA samples with the peak ratios of both alleles of a series of matched normal control samples. The peak ratio difference between control samples and tumor samples was considered to be an allelic imbalance if the peak ratio of the tumor sample was 50% higher or lower than the peak ratio of the control samples. We used 62 normal control DNA samples. All tumor samples were processed and analyzed in duplicate.
RESULTS
Clinical Data
The relevant clinical data of the 13 patients are summarized in Table 1. The male to female ratio was 0.63. The median age was 56 years, ranging from 46 to 78 years. Bone marrow involvement was detected in 10 patients. In 10 patients, tumor cells could be detected in the peripheral blood by morphology. Treatment consisted of splenectomy without additional therapy in 6 patients. In 7 patients, splenectomy was performed with subsequent chemotherapy. Transformation to a diffuse large B-cell lymphoma during follow-up was observed in four patients (Cases 5, 7, 9, 11). Five patients died, including all four patients with transformation to large-cell lymphoma. Median survival was not reached after a median follow-up of 5 years.
The immunophenotype in tissue and peripheral blood of all cases is summarized in Table 2. The CGH and cytogenetic results are summarized in Table 3. CGH revealed DNA copy number changes in 11 of 13 cases (85%). In 1 case, 27 abnormalities were found with CGH. In Case 8, we identified a deletion of chromosome 7q31–q34 as the only abnormality by cytogenetics as well as CGH. The most frequent changes found (summarized in Table 4) were gains of chromosome X (4 cases), chromosome 3 (3 cases), and chromosome 18 (3 cases) and losses of chromosome 7 (4 cases) and chromosome 6 (2 cases). In one of four cases with gains of chromosome X, a trisomy X was detected, and in two of three cases with gains of chromosome 3, a trisomy 3 was found; the other cases showed gain of parts of chromosome X and chromosome 3. Deletions of chromosome 17p11 were found in Cases 5 and 13. All three cases with gains on chromosome 18 had trisomy 18. Overall, chromosomal gains (28 ×) were more frequently found than losses (21 ×). In none of the cases was a high-level amplification (ratio of >2) detected. Strong overrepresentation (ratio of >1.5) was found in two patients for chromosome X.
In 10 of 13 cases we obtained cytogenetic data (see Table 3). Cytogenetic data were confirmed by the CGH results. In 9 cases the cytogenetic data demonstrated clonal abnormalities, frequently consisting of different subclones.
In 11 of 13 patients, atypical cells were observed in the peripheral blood by morphology, flow cytometry, or both (see Table 2). A loss of 7q was found with CGH and cytogenetics in three splenic cases with, and in one splenic case without, peripheral blood involvement. A ratio profile of the four cases with loss of 7q is shown in Figure 1.
The LOH results are summarized in Table 5, and an example of a case with an allelic imbalance of 7q31–32 is shown in Figure 2. Allelic imbalance was detected in 9 of 11 informative cases. The two exceptions were Case 9 and Case 12, which were the only cases with dim CD23 expression. Case 10 is of special interest because it showed a translocation (3;7)(q27;q32), and in the LOH assay an allelic imbalance of 7q31–32 was present. Also, Cases 1 and 11, which lacked chromosomal or CGH abnormalities, were found to have allelic imbalances of 7q31–32 by LOH.
Comparison of normal sample and tumor sample (Case 6) by LOH demonstrating allelic imbalance (AI) at 7q31. Samples were amplified for the D7S686 locus using fluorescent primers. Relative fluorescence is plotted against the size of the PCR product. The allele ratios are given in the right corner of each panel.
None of the cases showed translocations (11;18) or (1;14) as frequently encountered in marginal zone lymphomas of MALT, and also no cases with translocations (11;14) or (14;18) were found in this series of SMZL. A summary of gains and losses by CGH, the results of LOH analysis of 7q31–32, and the presence or absence of transformation to large cell lymphoma and of peripheral blood involvement are shown in Table 4.
DISCUSSION
In the present study we analyzed 13 cases of SMZL to identify chromosomal abnormalities using CGH and cytogenetics. CGH is a technique that analyzes DNA copy number losses and gains across a genome (10). In 11 of these 13 cases, abnormalities were found by CGH (86%), whereas in 2 cases no abnormalities were detected with CGH. In one of those two cases (Case 1), a loss of chromosome 19 was present in 4/24 metaphases with cytogenetics. In 8 of the 11 cases with abnormalities found by CGH, we also detected abnormalities with cytogenetics (see Table 3 for results). In most cases the cytogenetic data were comparable with the CGH data.
There are limited cytogenetic data available in the literature on SMZL. Some groups have performed cytogenetic analysis with the use of G-banding, FISH, and LOH, but only few cases of SMZL have been analyzed with CGH (5, 6, 8, 10, 14, 15, 16).
The most frequent abnormalities found in this study were gains in chromosomes 3 (23%), 18, (23%) and X (31%) and deletions in chromosomes 7q (31%) and 6q (15%; see Table 4 for summary). In four cases we found gain of chromosome X or parts of chromosome X. Gain of chromosome X was found in both female and male patients. Presence of an extra copy of chromosome X is commonly found as a secondary abnormality in NHL and probably is not important in the pathogenesis of NHL (17), although it may result in a growth advantage in some lymphomas (18).
In three cases we found abnormalities of chromosome 3, including trisomies of chromosome 3 in two cases. Previous cytogenetic studies on SMZL have shown variable percentages of trisomy 3, which range from 18 to 54% of the cases (6, 7, 14, 19). Although trisomy 3 is frequently found in marginal zone lymphomas of MALT, its exact role in the pathogenesis of SMZL is unknown. Recently it was proposed that there might be two forms of SMZL, one with gain of 3q and one with deletions at 7q; only one case in that study had a gain in 3q and a deletion in 7q (9). In our study, none of the cases showed a combination of these two aberrations with CGH and cytogenetics; however, two cases with gain in 3q had allelic imbalance of 7q31–32 in the LOH assay. There were two cases with translocations involving 3q27, the chromosomal region of BCL-6. In Case 5, 3q27 was involved in a t(3;17)(q27–29;q11–21), and in Case 8, it was involved in a t(3;7)(q27;q32).
We found trisomy 18 in three cases. Involvement of 18q21 is often associated with the t(11;18)(q21;q21) translocation; this translocation seems to be restricted to marginal zone lymphomas of MALT and has not been reported in SMZL (20, 21). Strong overrepresentation of 18q21–23 in marginal zone lymphoma was reported to be associated with BCL-2 gene overexpression (8). We did not detect strong overrepresentation of chromosome 18.
In two cases we found a deletion in 6q. In both cases, chromosome region 6q24 was involved. This included one of the four cases with secondary transformation to large cell lymphoma. Del 6q23–24 is among the most frequently occurring secondary chromosomal abnormalities that is found in non-Hodgkin’s lymphoma (17, 22). Several studies have identified possible hotspots on chromosome 6q. Three minimal molecular deletion regions (RMD) were detected. RMD1 located on chromosome 6q25-q27 was associated with intermediate-grade NHL, RMD2 located at 6q21 was associated with high-grade NHL, and RMD3 located at 6q23 was associated with low-grade NHL with and without t(14;18). All three RMD areas are suspected to harbor tumor suppressor genes (23).
In only one case (see Table 4) was gain of chromosome 18 found together with gain of chromosome 3. In two cases, we found a deletion of 6q24 in combination with a deletion of 7q31. The significance of these combinations is unknown. Comparing the abnormalities that are frequently found in chromosomes X, 3, and 18 and in chromosome 6q with clinical data like age, sex, bone marrow involvement, leukemic picture, stage, response to therapy, and survival did not reveal any obvious correlation. Four cases (Cases 5, 7, 9, and 11) underwent secondary transformation to a large-cell lymphoma. No correlation with any of the abnormalities in the primary lymphoma was found. However, the transformed cases included four of the five deceased patients.
By using CGH, we found a deletion of chromosome 7q ranging from 7q31 to 7q35 in four cases. In Case 3 (see Fig. 1), the ratio profile line is located on the loss borderline, whereas cytogenetic results showed a deletion of chromosome 7q21–32. This borderline loss is probably caused by the high percentage of normal cells in the tumor sample, which leads to averaging of the ratio profile. Del 7q is an abnormality that is often found in SMZL, SLVL, acute myeloid leukemia (AML), myelodysplastic syndromes (MDS), and prostatic cancer (8, 9, 12, 24, 25). In MDL, AML, and prostatic cancer, a correlation of the occurrence of del 7q32 and tumor progression has been reported (24, 26). A deletion of 7q was reported in three of five cases of a possible aggressive variant of SMZL, and it was suggested that a deletion of 7q31–32 is a marker for a more aggressive behavior of SMZL (26). In a recent study, approximately half of the cases of SMZL were found to have unmutated IgVH genes, and these cases were found to have relatively frequent 7q deletions and an adverse clinical course (27). In our study, no correlation was found between the occurrence of loss of 7q31–32 and a more aggressive behavior.
A microsatellite analysis was performed in all cases to determine the presence of allelic imbalances at 7q31–32 in cases that were not detected or that were not detectable because of the small size of the deletion by CGH and cytogenetics. LOH of 7q31–32 was detected in 9 of 11 informative cases. CGH and cytogenetics detected loss of 7q31 in four cases that also showed allelic imbalance for at least one of the three microsatellite markers. It is of interest that the two cases with a slightly aberrant immunophenotype, Cases 9 and 12, which had CD23 expression on part of the cells in the spleen as well as in the circulating cells, were the only two cases with no allelic imbalance. The two cases that were not informative (Cases 5 and 13) were the only two cases with a 17p deletion. Deletions in 17p that may include p53 are associated with progressive disease and poor response to therapy in several types of NHL (28). Case 5 was indeed one of the four cases that transformed to a large-cell lymphoma. Case 13 had extensive lymph node involvement in addition to splenomegaly. In the literature, a frequency of 10 to 15% has been reported for p53 abnormalities in SMZL or SLVL. Gruszka-Westwood et al. (29) found abnormalities in approximately 15% of SLVL cases, and these had a more aggressive disease course and prognosis. Sol Mateo et al. (30) investigated p53 mutations in 20 SMZL patients and found p53 mutations in 2 cases. Camacho et al. (31) described 12 cases of SMZL transforming to large B-cell lymphoma and detected p53 inactivation in only 1 case. The low frequency of 17p deletions observed in our transformed SMZL cases is in concordance with these results. On the other hand, Baldini et al. (32) found p53 mutations in 6 of 15 (40%) nonvillous SMZL cases and concluded that this was associated with poor prognosis. However, this study from 1994 included 7/15 cases with CD5 and/or CD23 expression, and of the 6 cases with p53 mutations, 4/6 had CD5 and/or CD23 expression. These cases would not have been included in more recent series of SMZL (29, 30, 31).
The nature of the putative suppressor gene involved in the 7q deletion is not known. Several studies have tried to narrow the common deleted region (CDR) on chromosome arm 7q. Mateo et al. (12) located the highest incidence of LOH at microsatellite D7S487, located at chromosome region 7q31–32, and Gruszka-Westwood et al. (33) found the CDR to be in chromosome region 7q32–33. It is now clear that it is not the CDK6 gene that is located at 7q21 and was previously thought to be involved in the pathogenesis of SMZL (34). It is of interest, however, that 7q31–35 deletion as the only chromosomal abnormality, as in Case 8, or 7q31 allelic imbalance, as in Cases 1 and 2, which had normal karyotypes by CGH and cytogenetics, are apparently sufficient to cause the proliferation of monoclonal marginal zone cells. Also of interest is the finding of a t(3;7)(q27;q32) in Case 10 that involves 3q27, the region of bcl6 and 7q32 as breakpoints.
Splenic lymphomas with CD5 or strong CD23 positivity lacked 7q31–32 deletion and/or allelic imbalance and were excluded from this study. The two cases with partial CD23 positivity that were included also lacked 7q31–32 deletion and/or imbalance. It can be concluded that SMZL presenting with prominent splenomegaly and the characteristic CD5−, CD10−, CD23− IgM+, IgDdim immunophenotype is a separate entity that is characterized genetically by 7q31–32 allelic imbalance.
References
Schmid C, Kirkham N, Diss T, Isaacson PG . Splenic marginal zone cell lymphoma. Am J Surg Pathol 1992; 16: 455–466.
Hollema H, Visser L, Poppema S . Small lymphocytic lymphomas with predominant splenomegaly: a comparison of immunophenotypes with cases of predominant lymphadenopathy. Mod Pathol 1991; 4: 712–717.
Pittaluga S, Verhoef G, Criel A, Wlodarska I, Dierlamm J, Mecucci C, et al. “Small” B-cell non-Hodgkin’s lymphomas with splenomegaly at presentation are either mantle cell lymphoma or marginal zone cell lymphoma. A study based on histology, cytology, immunohistochemistry, and cytogenetic analysis. Am J Surg Pathol 1996; 20: 211–223.
Hammer RD, Glick AD, Greer JP, Collins RD, Cousar JB . Splenic marginal zone lymphoma. A distinct B-cell neoplasm. Am J Surg Pathol 1996; 20: 613–626.
Brynes RK, Almaguer PD, Leathery KE, McCourty A, Arber DA, Medeiros LJ, et al. Numerical cytogenetic abnormalities of chromosomes 3, 7, and 12 in marginal zone B-cell lymphomas. Mod Pathol 1996; 9: 995–1000.
Dierlamm J, Pittaluga S, Wlodarska I, Stul M, Thomas J, Boogaerts M, et al. Marginal zone B-cell lymphomas of different sites share similar cytogenetic and morphologic features. Blood 1996; 87: 299–307.
Sole F, Salido M, Espinet B, Garcia JL, Martinez Climent JA, Granada I, et al. Splenic marginal zone B-cell lymphomas: two cytogenetic subtypes, one with gain of 3q and the other with loss of 7q. Haematologica 2001; 86: 71–77.
Dierlamm J, Rosenberg C, Stul M, Pittaluga S, Wlodarska I, Michaux L, et al. Characteristic pattern of chromosomal gains and losses in marginal zone B cell lymphoma detected by comparative genomic hybridization. Leukemia 1997; 11: 747–758.
Hernandez JM, Garcia JL, Gutierrez NC, Mollejo M, Martinez-Climent JA, Flores T, et al. Novel genomic imbalances in B-cell splenic marginal zone lymphomas revealed by comparative genomic hybridization and cytogenetics. Am J Pathol 2001; 158: 1843–1850.
Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992; 258: 818–821.
Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, et al. The 1993–94 Genethon human genetic linkage map. Nat Genet 1994;1993: 7: 246–339.
Mateo M, Mollejo M, Villuendas R, Algara P, Sanchez-Beato M, Martinez P, et al. 7q31–32 allelic loss is a frequent finding in splenic marginal zone lymphoma. Am J Pathol 1999; 154: 1583–1589.
Cuneo A, Bigoni R, Roberti MG, Moretti S, Bigoni R, Roberti MG, et al. Molecular cytogenetic characterization of marginal zone B-cell lymphoma: correlation with clinicopathologic findings in 14 cases. Haematologica 2001; 86: 64–70.
Sole F, Woessner S, Florensa L, Espinet B, Mollejo M, Martin P, et al. Frequent involvement of chromosomes 1, 3, 7 and 8 in splenic marginal zone B-cell lymphoma. Br J Haematol 1997; 98: 446–449.
Dierlamm J, Wlodarska I, Michaux L, Stefanova M, Hinz K, Van Den Berghe H, et al. Genetic abnormalities in marginal zone B-cell lymphoma. Hematol Oncol 2000; 18: 1–13.
Dargent JL, Delville JP, Kornreich A, Pradier O, Cochaux P, Velu T, et al. Morphologic and phenotypic changes of the leukemic cells in a case of marginal zone B-cell lymphoma. Ann Hematol 1997; 74: 149–153.
Johansson B, Mertens F, Mitelman F . Cytogenetic evolution patterns in non-Hodgkin’s lymphoma. Blood 1995; 86: 3905–3914.
McDonald HL, Gascoyne RD, Horsman D, Brown CJ . Involvement of the X chromosome in non-Hodgkin lymphoma. Genes Chromosomes Cancer 2000; 28: 246–257.
Wotherspoon AC, Finn TM, Isaacson PG . Trisomy 3 in low-grade B-cell lymphomas of mucosa-associated lymphoid tissue. Blood 1995; 85: 2000–2004.
Remstein ED, James CD, Kurtin PJ . Incidence and subtype specificity of API2-MALT1 fusion translocations in extranodal, nodal, and splenic marginal zone lymphomas. Am J Pathol 2000; 156: 1183–1188.
Rosenwald A, Ott G, Stilgenbauer S, Bredt M, Katzenberger T, Greiner A, et al. Exclusive detection of the t(11;18)(q21;q21) in extranodal marginal zone B cell lymphomas (MZBL) of MALT type in contrast to other MZBL and extranodal large B cell lymphomas. Am J Pathol 1999; 155: 1817–1821.
Zhang Y, Weber MK, Siebert R, Matthiesen P, Schlegelberger B . Frequent deletions of 6q23–24 in B-cell non-Hodgkin’s lymphomas detected by fluorescence in situ hybridization. Genes Chromosomes Cancer 1997; 18: 310–313.
Offit K, Parsa NZ, Gaidano G, Filippa DA, Louie D, Pan D, et al. 6q deletions define distinct clinico-pathologic subsets of non-Hodgkin’s lymphoma. Blood 1993; 82: 2157–2162.
Velloso ER, Michaux L, Ferrant A, Hernandez JM, Meeus P, Dierlamm J, et al. Deletions of the long arm of chromosome 7 in myeloid disorders: loss of band 7q32 implies worst prognosis. Br J Haematol 1996; 92: 574–581.
Takahashi S, Shan AL, Ritland SR, Delacey KA, Bostwick DG, Lieber MM, et al. Frequent loss of heterozygosity at 7q31.1 in primary prostate cancer is associated with tumor aggressiveness and progression. Cancer Res 1995; 55: 4114–4119.
Lloret E, Mollejo M, Mateo MS, Villuendas R, Algara P, Martinez P, et al. Splenic marginal zone lymphoma with increased number of blasts: an aggressive variant? Hum Pathol 1999; 30: 1153–1160.
Algara P, Mateo MS, Sanchez-Beato M, Mollejo M, Navas IC, Romero L, et al. Analysis of the IgV(H) somatic mutations in splenic marginal zone lymphoma defines a group of unmutated cases with frequent 7q deletion and adverse clinical course. Blood 2002; 99: 1299–1304.
Koduru PR, Raju K, Vadmal V, Menezes G, Shah S, Susin M, et al. Correlation between mutation in P53, p53 expression, cytogenetics, histologic type, and survival in patients with B-cell non-Hodgkin’s lymphoma. Blood 1997; 90: 4078–4091.
Gruszka-Westwood AM, Hamoudi RA, Matutes E, Tuset E, Catovsky D . p53 abnormalities in splenic lymphoma with villous lymphocytes. Blood 2001; 97: 3552–3558.
Sol Mateo M, Mollejo M, Villuendas R, Algara P, Sanchez-Beato M, Martinez-Delgado B, et al. Analysis of the frequency of microsatellite instability and p53 gene mutation in splenic marginal zone and MALT lymphomas. Mol Pathol 1998; 51: 262–267.
Camacho FI, Mollejo M, Mateo MS, Algara P, Navas C, Hernandez JM, et al. Progression to large B-cell lymphoma in splenic marginal zone lymphoma: a description of a series of 12 cases. Am J Surg Pathol 2001; 25: 1268–1276.
Baldini L, Guffanti A, Cro L, Fracchiolla NS, Colombi M, Motta M, et al. Poor prognosis in non-villous splenic marginal zone cell lymphoma is associated with p53 mutations. Br J Haematol 1997; 99: 375–378.
Gruszka-Westwood AM, Hamoudi R, Osborne L, Matutes E, Catovsky D . Deletion mapping on the long arm of chromosome 7 in splenic lymphoma with villous lymphocytes. Genes Chromosomes Cancer 2003; 36: 57–69.
Corcoran MM, Mould SJ, Orchard JA, Ibbotson RE, Chapman RM, Boright AP, et al. Dysregulation of cyclin dependent kinase 6 expression in splenic marginal zone lymphoma through chromosome 7q translocations. Oncogene 1999; 18: 6271–6277.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Boonstra, R., Bosga-Bouwer, A., van Imhoff, G. et al. Splenic Marginal Zone Lymphomas Presenting with Splenomegaly and Typical Immunophenotype Are Characterized by Allelic Loss in 7q31–32. Mod Pathol 16, 1210–1217 (2003). https://doi.org/10.1097/01.MP.0000095895.19756.77
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.MP.0000095895.19756.77
Keywords
This article is cited by
-
Mutational landscape of marginal zone B-cell lymphomas of various origin: organotypic alterations and diagnostic potential for assignment of organ origin
Virchows Archiv (2022)
-
The prevalence of IG translocations and 7q32 deletions in splenic marginal zone lymphoma
Leukemia (2008)
-
A comprehensive genetic and histopathologic analysis identifies two subgroups of B-cell malignancies carrying a t(14;19)(q32;q13) or variant BCL3-translocation
Leukemia (2007)
-
Cytogenetic analysis delineates a spectrum of chromosomal changes that can distinguish non-MALT marginal zone B-cell lymphomas among mature B-cell entities: a description of 103 cases
Leukemia (2005)