Abstract
Differentiation between well-differentiated hepatocellular carcinoma (HCC) and nonmalignant lesions with increased cellular proliferation may be difficult in needle biopsies. Based on recurrent chromosome aberrations known for HCC, we developed a nonfluorescent in situ hybridization technique that allows combination with morphological analysis in bright-field microscopy. Fourteen biopsies of HCC and 31 samples of regenerative nodules (n = 10), chronic hepatitis (n = 10), fibrosis or cirrhosis of unknown origin (n = 5), focal nodular hyperplasia (n = 2), primary biliary cirrhosis (n = 2), steatosis (n = 1), and adenomatous hyperplasia (n = 1) were analyzed with probes specific for the centromeric regions of chromosomes 1, 6, 7, and 8. After microwave pretreatment and in situ hybridization, signals were detected using a tyramine-based system and AEC as substrate. Evaluation of signals was done by conventional bright-field microscopy. Using this approach, aberrant counts were seen for at least one chromosome in 12/14 cases of HCC. In contrast, none of the nonmalignant lesions revealed aberrant counts for any of the chromosomes analyzed. In conclusion, this new combination of in situ hybridization and tyramine amplification allows fast and reliable evaluation of chromosome aberrations in a histomorphological context similar to paraffin immunohistochemistry. Registration of imbalances contributes to a reliable differentiation between malignant and nonmalignant lesions of the liver.
Similar content being viewed by others
INTRODUCTION
The fast and reliable differentiation between malignant and nonmalignant tumorlike lesions of the liver is of the utmost importance for the further treatment and surgical procedure of the patients. However, even for the experienced pathologist, in some cases, determining the correct diagnosis may be extremely difficult and uncertain, particularly if only small biopsies are available as the main source for histological examination. In particular, differentiation between well-differentiated hepatocellular carcinoma (HCC) and benign alterations may be impossible based on morphologic criteria alone (1).
Detection of chromosomal aberrations within questionable tumors could contribute toward solving this problem. Because of the time and effort required for conventional cytogenetics, this technique is usually not appropriate. The recently developed comparative genomic hybridization (CGH) has revealed promising results (2, 4, 11, 13, 14), but it is also of limited value because of the duration and amount of tissue required. By contrast, fluorescence in situ hybridization (FISH) yields results that are evaluable by simple counting of fluorescence signals in conventional biopsies (12). A major limitation of this approach is, however, that most pathologists are not familiar with the morphology of the tumor in histological sections counterstained with fluorescent dyes. Furthermore, many histomorphological details remain undetected in the dark field required for evaluation. In addition, epifluorescence microscopy is based on expensive technical equipment.
In this setting, we describe a new way to circumvent these limitations. A new combination of in situ hybridization (ISH) and tyramine signal amplification was established for the detection of chromosomal imbalances in liver tumors. It enables the determination of chromosomal imbalances by conventional bright-field microscopy. We applied this technique to 45 biopsy specimens of malignant and nonmalignant tumor-like lesions in a blind study.
MATERIAL AND METHODS
Forty-five specimens of patients at Hannover Medical School, Hannover, Germany were examined in this study. Age, gender, and histological diagnosis are given in Table 1. Histological diagnoses were obtained using hematoxylin and eosin–stained sections as well as sections stained with periodic acid–Schiff, Elastica van Gieson, silver, and Prussian blue. In 14 cases, well-differentiated HCC was seen, whereas in the remaining 31 patients, nonneoplastic alterations were detected, in particular, regenerative nodules (n = 10), chronic hepatitis (n = 10), fibrosis or cirrhosis of unknown origin (n = 5), steatosis (n = 1), focal nodular hyperplasia (n = 2), primary biliary cirrhosis (n = 2), and adenomatous hyperplasia (n = 1). As controls, 30 specimens of normal livers were used.
In Situ Hybridization
ISH for centromeric regions of chromosomes 1, 6, 7, and 8 (D1Z1, D6Z1, D7Z1, D8Z2) was performed for all samples using probes purchased from Oncor (Heidelberg, Germany). Thickness of sections was set at 5 μm based on our own experience (12) and the reports of other authors (8). This value was a compromise with regard to an adequately preserved morphology and the effect of nuclei cutting leading to a diminishing of the number of signals. Tissues were mounted on Superfrost Plus slides (Omnilab, Hannover, Germany).
Tissue sections were baked overnight at 56° C and then deparaffinized by immersion in xylene for 20 minutes and in graded ethanol. Slides were then bathed in citric acid solution (6 m) and heated in a microwave oven at 900 and 600 W for 15 minutes each. RNase A (0.1%) was added to the sections for 10 minutes and then rinsed in PBD (Oncor, Heidelberg, Germany). Incubation with 3% H2O2 for 10 minutes at room temperature followed and was stopped by washing in PBD. Afterwards, slides were washed in graded ethanol and air dried for 5 minutes. A half microliter of the probe was added to 10 μL of Hybrisol VI (Oncor, Heidelberg, Germany) and pipetted onto the slide, covered with a coverslip, sealed with rubber cement, heated up to 92° C for 12 minutes, and incubated overnight at 37° C in a humidified chamber. Detection commenced with rinsing in 0.25× standard saline citrate at 60° C for 5 minutes, followed by a short wash in PBD. Then, 30 μL horseradish peroxidase (HRP), diluted 1:30, were added for 20 minutes at 37° C under a coverslip. Washing in PBD followed. 30 μL of biotin-conjugated tyramine (DuPont NEN, Boston, MA) were added, and the mixture was incubated for 20 minutes at 37° C. After rinsing in PBD, incubation with anti-biotin horseradish peroxidase, diluted 1:30 in water, followed for 20 minutes under a coverslip at room temperature. Rinsing was performed again in PBD, followed by incubation for 10 minutes at room temperature with AEC+/substrate Chromogen (DAKO, Hamburg, Germany). Specimens were rinsed in deionized water. Counterstaining was carried out with hemalaun for a few seconds followed by rinsing in water. Glycerin gelatin was added, and a coverslip was laid on the tissue.
Signals were evaluated using a standard microscope (Axiophot, Zeiss, Oberkochen, Germany). Only clearly detectable signals not connected to a second signal were counted. For each case, ≥100 nuclei were evaluated.
RESULTS
Control Group
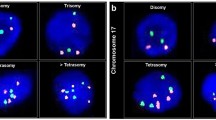
In these specimens, depending on the probe used, 84–87% of the nuclei displayed two signals, 11–13% revealed one signal, and 2–3% showed three or more signals. The standard deviations were 4.5–5.8%, 4.7–5.3%, and 0.9–1.7%, respectively. Regarding the recommendations of Ward et al. (10), three standard deviations were added to the mean values and defined monosomy for centromeric probes 1, 6, 7, and 8 at 26, 27, 30, and 25%, respectively. Trisomy was defined at values of 5, 6, 7, and 6% (data not shown).
Nonneoplastic Specimens
In the 31 nonneoplastic biopsy specimens analyzed, the mean value for two signals per nucleus was 87% (Table 2, Fig. 1), 12% revealed one signal, and 1% of nuclei showed three or more signals. The standard deviations were 5.3%, 5.4%, and 1.0%, respectively. In none of these specimens was the diagnostic threshold for monosomy or trisomy reached.
Well-Differentiated HCC
Evaluation ofHCC revealed very different results. In these samples (Table 3, Fig. 2), one signal was seen in 4.2 to 66.9% of cells (mean: 18.2%, standard deviation: 14.1%). Two signals were seen in 32.8 to 92.3% (mean: 69.6%, standard deviation: 16.4%), and three signals occurred in 0 to 62.8% (mean: 12.2%, standard deviation: 16.6%). Chromosome 1 was found to be aberrant in 11 samples; chromosome 6, in 6 samples; chromosome 7, in 5 samples; and chromosome 8, in 7 samples. Four chromosomes were found aberrant in 2 samples, 3 chromosomes were aberrant in 4 samples, 2 chromosomes were aberrant in 4 samples, and only 1 chromosome was found aberrant in 2 samples. Two samples revealed no aberrant counts at all (Table 3).
Statistical Evaluation
The number of aberrations found in nonmalignant lesions and HCC samples was statistically different, with high significance (P < .01, Mann-Whitney U test). Specificity of ISH in detecting HCC was 100%, and sensitivity was 80%. Positive predictive value was 100%, and the negative predictive value reached 91%.
DISCUSSION
The new technique described in this study employs results obtained by CGH (4, 13, 14). Even in well-differentiated HCC, numerous recurrent aberrations have been found (2). In contrast, benign tumors such as hepatocellular adenomas did not reveal these chromosome alterations (11). Based on these data, we used a combination of in situ hybridization and immunohistochemistry to detect these aberrations in a simple way. The results obtained by this approach allowed the correct detection of well-differentiated HCC in 80% of cases. The correct negative predictive value reached 91%. Most important, in none of the samples analyzed was a false-positive result found. These findings in well-differentiated HCC are very similar to those found in an earlier study of our group using the FISH technique (12). In that study, aberrant counts were detected in 13/14 cases. The hepatocellular adenomas analyzed in parallel revealed no aberrations in any samples for probes specific for centromeres of 1, 6, 7, and 8. This agrees with the results on the nonmalignant lesions of this study. Although these alterations of liver architecture are assumed to be associated with an increased rate of cell proliferation, a normal distribution of signals was found in all samples examined. In particular, percentages of nuclei bearing more than two signals in the nonmalignant lesions differ significantly from HCC.
However, normal counts were also detected in 2/14 HCC samples analyzed and in 1/13 HCC examined in the FISH study (12). This was also seen in the CGH studies mentioned above, supporting the assumption that chromosomal aberrations probably occur as later steps in the development of HCC at least in some of the tumors. The assumption is also emphasized by the observation that these aberrations are also found in a variety of other neoplasms as well-known changes (5). Some investigators have discussed them as a consequence of genetic instability caused by mutations at an earlier level (3). The basic events leading to these findings are still not defined and require further, more sophisticated evaluation.
As reported repeatedly, FISH is more sensitive than ISH (7). Earlier attempts to improve ISH using immunohistochemistry approaches instead of radioactivity were hampered by either low sensitivity or background staining, making it difficult to evaluate signals. These problems have now been circumvented by changes in the experimental set-up reported here and in variant forms by other investigators, too (7, 9). Differences in these experimental approaches are found mainly in the pretreatment steps and in the method of detection of the hybridized nucleic acids. Vos et al. (9) used proteolytic enzyme digestion as pretreatment, with the disadvantage of testing an optimized digestion time. Tanner et al. (7) preferred a combination of enzymatic digestion and microwave heating and found that this testing was no longer necessary. This was also seen in our study using microwave heating alone. Signal detection was performed by Vos et al. (9), with tyramine amplification similar to our protocol. Tanner et al. (7) used repeated incubations with labeled antibodies to enhance signal intensity.
The signals found with the combination of techniques used in this study are easy to count and differ clearly from the spots seen as background staining in the cells examined. The evaluation of signals is even more simple than in FISH because hybridized biopsies can be examined using simple, standard microscopes. Moreover, counting of signals by ISH is made easier by counterstaining cells with hemalaun, a stain well known in immunohistochemistry. In many samples, the strong signals and the standard counterstaining give an excellent overview, allowing a first impression of the results and a very fast detailed evaluation of specimens. However, it has to be kept in mind that evaluation of signals is hampered in general by cutting artifacts based on truncation of nuclei. This effect leads to a decrease in signal number, with two signals in approximately 80–85% of the nuclei instead of 95% normally found in preparations of intact nuclei (6, 8, 15). In particular, detection of monosomy may be difficult, with thresholds at nearly 30%. Therefore, to exclude monosomy based on low hybridization efficiency leading to false-positive results, we recommend the additional evaluation of an internal standard, such as nonneoplastic cells situated near or in the tumor.
In conclusion, this ISH technique yields reliable results and can contribute to the differentiation between well-differentiated HCC and nonmalignant lesions. Results are obtained within 24 hours with minimum effort and require only standard microscope techniques.
References
Edmondson HA, Steiner PE . Primary carcinoma of the liver; a study of 100 cases among 48900 necropsies. Cancer 1954; 7: 462–503.
Kusano N, Shirashi K, Kubo K, Oga A, Okita K, Sasaki K . Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology 1999; 29: 1858–1862.
Lengauer CL, Kinzler KW, Vogelstein B . Genetic instabilities in human cancer. Nature 1998; 396: 643–649.
Marchio A, Meddeb M, Pineau P, Danglot G, Tiollais P, Bernheim A, et al. Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosom Cancer 1997; 18: 59–65.
Mitelman F, Mertens F, Johansson B . A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat Genet 1997; 15(Spec No): 417–474.
Slagel DD, Bromley CM, Benda JA . Detection of chromosomal abnormalities in the dysmorphic fetus using fluorescence in situ hybridization: evaluation for monosomy X genotype. Hum Pathol 1995; 26: 1241–1244.
Tanner MM, Gancberg D, Di Leo A, Larsimont D, Rouas G, Piccart MJ, et al. Chromogenic in situ hybridization: a practical alternative for fluorescence in situ hybridization to detect Her-2/neu oncogene amplification in archival breast cancer samples. Am J Pathol 2000; 157: 1467–1472.
Visscher DW, Wallis TL, Crissman JD . Evaluation of chromosome aneuploidy in tissue sections of preinvasive breast carcinomas using interphase cytogenetics. Cancer 1996; 77: 315–320.
Vos CBJ, Ter Haar NT, Peterse JL, Cornelisse CJ, van de Vijver MJ . Cyclin D1 gene amplification and overexpression are present in ductal carcinoma in situ of the breast. J Pathol 1999; 187: 279–284.
Ward BE, Gersen SL, Carelli MP, McGuire NM, Dackowski WR, Weinstein M, et al. Rapid prenatal diagnosis of chromosomal aneuploidies by fluorescence in situ hybridization: clinical experience with 4,500 specimens. Am J Hum Genet 1993; 52: 854–865.
Wilkens L, Bredt M, Flemming P, Becker T, Kubicka S, Kreipe H . Differentiation of liver cell adenomas from well differentiated hepatocellular carcinomas by comparative genomic hybridisation. J Pathol 2001; 193: 476–482.
Wilkens L, Bredt M, Flemming P, Schwarze Y, Becker T, Klempnauer J, et al. Diagnostic impact of fluorescence in situ hybridisation in the differentiation of hepatocellular adenoma and well differentiated hepatocellular carcinoma. J Mol Diagn 2001; 3: 68–73.
Wong N, Lai P, Lee SW, Fan S, Pang E, Liew CT, et al. Assessment of genetic changes in hepatocellular carcinoma by comparative genomic hybridization analysis. Am J Pathol 1999; 154: 37–43.
Zimonjic DB, Keck CL, Thorgeirsson SS, Popescu N . Novel recurrent genetic imbalances in human hepatocellular carcinoma cell lines identified by comparative genomic hybridization. Hepatology 1999; 29: 1208–1214.
Zitzelsberger H, Szucs S, Weier HU, Lehmann L, Braselmann H, Enders S, et al. Numerical abnormalities of chromosome 7 in human prostate cancer detected by fluorescence in situ hybridization (FISH) on paraffin-embedded tissue sections with centromere-specific DNA probes. J Pathol 1994; 172: 325–335.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wilkens, L., Bredt, M., Flemming, A. et al. Detection of Chromosomal Aberrations in Well-Differentiated Hepatocellular Carcinoma by Bright-Field In Situ Hybridization. Mod Pathol 15, 470–475 (2002). https://doi.org/10.1038/modpathol.3880548
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3880548
Keywords
This article is cited by
-
Image-based computational quantification and visualization of genetic alterations and tumour heterogeneity
Scientific Reports (2016)
-
Chromogenic In Situ Hybridization Analysis of Epidermal Growth Factor Receptor Gene/Chromosome 7 Numerical Aberrations in Hepatocellular Carcinoma Based on Tissue Microarrays
Pathology & Oncology Research (2009)
-
Leberzelladenom
Der Pathologe (2006)
-
Late-onset Epstein–Barr virus (EBV)-negative extranodal B-precursor lymphoblastic lymphoma of donor origin after hematopoietic stem cell transplantation (HSCT)
Bone Marrow Transplantation (2003)