Abstract
Mutations of the p53 gene are one of the most common genetic changes found in cancer; their presence may be prognostic and even influence treatment for breast cancer. In this study, we investigated whether DNA could be extracted from the residual cells left in ThinPrep-processed breast fine-needle aspirates and whether p53 gene changes could be detected in the DNA. The results were then correlated with DNA extracted from the matched formalin-fixed, paraffin-embedded, surgically resected breast cancer when available. DNA was successfully extracted from 54 of 62 aspirates and all 31 surgical specimens. p53 gene mutations were detected in 10 of the 54 cytology specimens (18.5%) and consisted of base pair substitutions or deletions. Silent or intronic p53 changes were found in five additional aspirates. One of the aspirates had two gene alterations, resulting in a total of six gene changes. Five of these changes were located in introns 6 or 9 and the sixth was a silent (no amino acid change) change in exon 6. p53 Polymorphisms were detected in nine aspirates (16.3%) and were located in codon 47 (one aspirate), codon 72 (six aspirates), and codon 213 (two aspirates). All cases with surgical material available showed identical p53 mutations, alterations, and polymorphisms in the resected tumors compared with those detected in the corresponding aspirates. The results of this study show that DNA suitable for analysis of p53 gene sequence changes can be successfully extracted from ThinPrep-processed breast fine-needle aspirates, and that identical alterations are detected in both the cytology and surgical specimens.
Similar content being viewed by others
INTRODUCTION
Mutations of the p53 gene are among the most common molecular changes detected in human cancers (1). Experimental studies have shown that functional p53 is required for the in vitro cytotoxic action of some chemotherapeutic agents (2) The presence of p53 mutations is associated with an increased chemoresistance to doxorubicin in breast cancer patients (3) and may be involved in the development of multidrug resistance (4). Clinical studies have shown that breast cancers that contain p53 gene mutations are associated with decreased disease-free and overall survival (3, 5, 6, 7, 8, 9). These results suggest that the presence of p53 mutations might provide prognostic information and influence the treatment of the breast cancer.
Fine-needle aspiration (FNA) of the breast is a safe, effective method for diagnosing breast cancer with minimal intervention and complications (10, 11). As reviewed by Bédard et al., for the detection of carcinoma, it has a sensitivity ranging from 74 to 97% and a specificity ranging from 82 to 100% (12). ThinPrep-processed and conventionally processed breast FNA have been shown to have similar diagnostic accuracy (12). In addition, immunohistochemistry (13, 14) and molecular analysis (15, 16, 17) have been successfully applied to ThinPrep-processed specimens.
Because FNA is often the initial sampling of the tumor, it could be a source of cells for the early detection of p53 mutations. In this study, we examined whether p53 mutations could be detected in the cells present in the residual fluid from ThinPrep-processed breast FNAs. When available, the corresponding paraffin-embedded surgically resected tissue was also analyzed for p53 mutations and the results were correlated.
MATERIALS AND METHODS
Specimen Acquisition, Clinical History, and Pathology Review
Cytology reports from November 1997 to April 1999 in the files of Mount Sinai Hospital were reviewed. Of the cases diagnosed as positive or suspicious for malignancy, DNA could be extracted from 54 of 62 specimens of ThinPrep processed breast FNA obtained from 62 different women. In cases in which DNA was successfully extracted from the cytology fluid, the surgical pathology records were reviewed to determine whether there was a corresponding breast tumor specimen. Formalin-fixed, paraffin-embedded tissue was available for 31 women. Clinical details and tumor characteristics were obtained from surgical reports. The breast cancers were graded according to the Elston’s modified Bloom and Richardson criteria (18). In 30 of the 31 surgical specimens, the tumor was removed after the cytology specimen. On average, the specimen was removed 33 days after the FNA (range, 8 to 72 days). In one case, the FNA was from a tumor recurrence in the scar 6 weeks after the mastectomy.
p53 Molecular Analysis
DNA Extraction: Cytology
After completing the cytological examination the residual preservative fluid (PreservCyt solution, Cytyc Corporation, Boxborough, MA) was stored at 4°C for up to 3 months. The fluid was centrifuged at 4000 g and the supernatant was removed. DNA was extracted from the remaining cells using TriZol (Gibco-BRL, Rockville, MD). DNA extraction was performed according to the manufacturer’s instructions for cells grown in suspension. The DNA was stored at 4°C until used for analysis.
DNA Extraction: Surgical Specimens
Sections (5 μm) were cut from the paraffin blocks and stored for up to 2 weeks. Before microdissection, the sections were dewaxed and stained briefly with hematoxylin. A representative portion of the tumor containing minimal numbers of stromal and inflammatory cells was microdissected and placed in a microfuge tube. The tissue was digested with proteinase K (0.5 mg/mL in 50 mm Tris·HCl, pH 8.5, 10 mm EDTA, 0.5% Tween 20) for at least 48 hours at 55°C (19). The proteinase K was inactivated by heating at 95°C for 15 minutes. The DNA was stored at −20°C for up to 3 wk until further analyzed.
Polymerase Chain Reaction (PCR)—Single Strand Conformational Polymorphism Analysis (SSCP)
A 1-μL aliquot from each sample was added to 14 μL of PCR solution containing 1.5 mm CaCl2, 20 mm Tris·HCl, pH 8.0, 50 mm KCl, 0.25 μm concentrations of each primer, 0.1 mm concentrations of each dNTP, 1 U Taq DNA polymerase (GibcoBRL, Rockville, MD), and 2 μCi [α-33P]dATP. The primers and the cycling conditions for each exon are listed in Table 1. The reaction product was run on an 8% nondenaturing polyacrylamide gel and the gel was processed for autoradiography (20, 21). Potential mutations were detected by shifts in band mobility. If there was no band shift, the tissue was considered to have no mutation. For samples showing band shifts, the PCR-SSCP analysis was repeated. In cases in which different band shifts were detected in the cytology and corresponding paraffin-embedded samples, an additional paraffin block was selected, cut, microdissected, and processed as above. Negative controls, paraffin-embedded cells that contained no p53 mutation in the exon examined and a water control to replace the DNA, were included in each analysis. Positive controls for exons 5 to 9 (exon 5, SKBr3; exon6, T47D; exon 7, colo 320DM; exon 8, MDA-MB468; exon 9, SW480) were also included where appropriate.
p53 Sequencing
The abnormally shifted band was excised from the SSCP gel and the DNA was eluted into water. The DNA was reamplified by PCR using the same primers and the product was run on a 2% agarose gel. The band was extracted using a QIAquick Gel Extraction Kit (QIAGEN, Chatsworth, CA). The purified DNA was sequenced using a ThermoSequenase radiolabeled terminator cycle sequencing kit (Amersham Life Sciences, Cleveland, Ohio) and the sense primer according to the manufacturer’s directions, followed by gel electrophoresis and autoradiography. To confirm the mutation, the DNA product was resequenced using the antisense primer. Negative controls were included in each analysis. Cell lines with known mutations in exons 5 to 9 were also included where appropriate. Mutations were compared with those mutations listed for breast cancer in a known p53 database (http://www.iarc.fr/p53) (22).
Statistical Analysis
The associations between p53 gene alterations and clinical/tumor variables were examined using the χ2 or, where appropriate, Fisher’s exact test (23). Two-sided P-values below 0.05 were considered to be statistically significant.
RESULTS
Histological review of the 31 surgically resected breast tumors showed that they consisted of 29 infiltrating ductal carcinomas not otherwise specified, one invasive ductal carcinoma with lobular features, and one mucinous carcinoma. DNA was successfully extracted from all paraffin-embedded tumors.
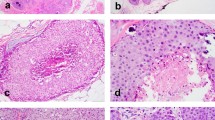
Of 62 cytology samples, DNA suitable for p53 sequencing was extracted from 54, yielding an evaluable specimen in 87% of the cases. p53 Gene mutations were detected in 10 of the 54 cytology specimens (18.5%). As shown in Table 2, these consisted of base pair substitutions and deletions. For eight of these 10 aspirates, surgically resected breast tumor tissue was available for gene analysis. All eight cases showed identical p53 mutations in both the aspirate and the surgically resected tumor. A representative SSCP gel is shown in Figure 1 and the associated sequencing gel is shown in Figure 1B.
A, a representative SSCP gel of p53 exon 7 PCR product from three cases and a negative control (Control). S-16 (surgically resected breast cancer) shows no abnormality. The cytology sample (C-38) and the corresponding paraffin-embedded surgical sample (S-17) show similar band shifts (→). B, the sequencing gel for samples C-38 and S-17 shows a T-to-G base substitution (→). The wild type sequencing pattern (control) in the same region is also shown.
Other types of p53 gene changes were found in five other aspirates. One aspirate had two gene alterations resulting in a total of six gene changes. As shown in table 3, five changes were located in introns 6 or 9 and one was a silent change (no amino acid change) in exon 6. For two of these five aspirates, surgically resected breast tumor tissue was available for gene analysis and both of the cases showed identical p53 gene changes in the aspirate and the surgically resected tumor.
p53 Polymorphisms were detected in nine aspirates (16.3%) and as shown in Table 4 were located in codon 47 (one aspirate), codon 72 (six aspirates), and codon 213 (two aspirates). For seven of these nine aspirates, surgically resected breast tumor tissue was available for gene analysis and all seven cases showed identical p53 polymorphisms in both the aspirate and the surgically resected tumor.
The clinical features and tumor characteristics were correlated with the p53 gene status and are summarized in Table 5. DNA suitable for p53 sequencing could be obtained from aspirates of tumors of all three grades. The women whose tumors had a p53 mutation or an intronic change or a silent change were grouped together for these analyses because of the small numbers. There was a significant correlation between a younger age (P = .038) or larger tumor size (P = .046) with the presence of p53 gene alterations. There was no correlation between the presence of estrogen (P = .449) or progesterone (P = 0.066) receptors or tumor grade (P = .227) and the presence of p53 gene alterations.
DISCUSSION
This study demonstrated that DNA can be extracted from ThinPrep processed breast FNAs. This is in keeping with the findings of other groups that have reported successful extraction of RNA or DNA from ThinPrep-processed cytology specimens of breast and cervix (15, 16, 17). In addition, the current study showed that the extracted DNA was suitable for p53 gene analysis by PCR-SSCP and sequencing. Using the protocol described above, the mutations detected in exons 4 to 9 were identical to those found in the formalin-fixed, paraffin-embedded, surgically resected breast cancer when this tissue was available for analysis. In contrast, studies assessing p53 immunoreactivity in FNAs and formalin-fixed, paraffin-embedded tumors have shown variable correlations ranging from 73.5 to 93.3% (24, 25, 26).
Recent studies have shown that gene alterations detected in paraffin-embedded tissue may be artifacts induced by fixation or processing of surgical specimens (27, 28). Several precautionary steps were undertaken to minimize this possibility. The fidelity of the PCR amplification of DNA extracted from paraffin can be markedly improved by prolonged proteinase K digestion and using small DNA templates (29), so in this study the paraffin-extracted DNA was digested by proteinase K for at least 48 h and the primers were chosen to provide gene sequences of less than 300 base pairs in length. To ensure that the gene alterations were not caused by nucleotide substitutions as a result of Taq DNA polymerase misincorporation, all specimens with abnormal SSCP underwent repeat PCR-SSCP to confirm that the change was reproducible. Only those samples that showed similar changes on the repeat PCR-SSCP were considered to have a sequence alteration, which was then confirmed by sequencing. Furthermore, identical alterations were seen in the methanol fixed aspirates and in the corresponding formalin-fixed, paraffin-embedded, surgically resected tumors. This suggests that the p53 alterations identified in this study were genuine.
p53 Mutations were found in 18.5% of patients. This is within the frequency reported for breast carcinoma in other series (8, 9, 30, 31, 32, 33, 34). The majority of changes reported for breast cancer have been point mutations (22), and in our series, eight of the 10 mutations (80%) involved base pair substitutions. All mutations, except two (cytology specimens 7 and 13) have been previously reported to occur in breast cancer as listed in a p53 database (22). Silent gene changes were detected in 1.9% of patients, which is similar to the frequency (1.8%) reported by Burns et al. (6). In the database examined, there was no report of the silent change observed at codon 224 (cytology specimen 56). No similar comparison could be done for the intronic alterations because the nucleotide position of these types of gene changes is not provided in the database. Codon 47 in exon 4, codon 72 in exon 4 and codon 213 in exon 6 contained known polymorphisms in one, six, and two patients, respectively (1.8, 11.1, and 3.7% of the patients). This is within the range determined for the normal population (35, 36, 37). Because the frequencies of mutations and polymorphisms are similar to those shown by others, this suggests that our methodology to detect p53 gene changes is appropriate.
The presence of p53 alterations showed statistically significant associations with larger tumors and younger patient age. No significant association was seen between p53 alterations and tumor grade or the presence or absence of estrogen and progesterone receptors. Other studies examining the association between these clinical variables and p53 protein accumulation and/or mutations have yielded inconsistent and often conflicting results. For example, Caleffi et al. found that p53 mutations occurred in younger patients (38) but other studies have not found an association between age and p53 status (5, 39, 40). The number of patients in the current report is small and may have compromised the statistical power of the study to detect associations.
The use of residual cells from ThinPrep-processed samples has several advantages. First, the fluid from ThinPrep processing can be stored at 4°C for up to 3 months, before extracting the DNA, as observed in the present study. Second, because only the residual fluid is needed for analysis, the original diagnostic slides do not have to be used. Third, in contrast to paraffin-embedded tissue, which has to undergo proteinase K digestion for at least 48 h before DNA extraction, ThinPrep-processed aspirates can undergo DNA extraction the same day they are obtained. However, there may also be disadvantages to using the residual material from ThinPrep-processing. Not all cases have tumor cells remaining in the residual fluid and thus DNA may not be available for analysis. In addition, if the aspirate contains numerous benign cells admixed with the malignant cells, mutations may be missed (20, 21).
Immunohistochemical staining can be used to detect p53 protein accumulation in either cytological or surgical specimens (24, 25, 26) but the immunohistochemical results do not always reflect the presence of underlying genetic changes (33, 34, 41, 42). For example, nonsense mutations will not cause protein accumulation, so these cells will be negative by immunohistochemical staining. In keeping with this, the presence of p53 mutations in the breast cancer was shown to be associated with decreased disease free survival as well as overall survival (5, 6, 7, 8, 9, 31), but the presence of p53 protein detected immunohistochemically in the tumor has not consistently been associated with a worse prognosis (7, 8, 42). As molecular analysis of p53 may provide prognostic and treatment information for patients with breast cancer, ThinPrep aspirate is a suitable alternative to the paraffin-embedded tissue as a source of cells for this type of analysis in patients who will receive neoadjuvant chemotherapy or have unresectable tumors.
In summary, ThinPrep-processed breast FNAs provide DNA suitable for molecular analysis more rapidly than paraffin-embedded tissue. FNAs seem to be a reliable source of cells to determine the p53 gene status, given that identical alterations were detected in both the cytology and surgical specimens examined in this study.
References
Levine AJ, Momand J, Finlay CA . The p53 tumour suppressor gene. Nature 1991; 351: 453–456.
Lowe SW, Ruley HE, Jacks T, Housman DE . p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 1993; 74: 957–967.
Ogretmen B, Safa AR . Expression of the mutated p53 tumor suppressor protein and its molecular and biochemical characterization in multidrug resistant MCF- 7/Adr human breast cancer cells. Oncogene 1997; 14: 499–506.
Aas T, Borresen AL, Geisler S, Smith-Sorensen B, Johnsen H, Varhaug JE, et al. Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med 1996; 2: 811–814.
Berns EMJJ, Klijn JG, Smid M, van Staveren IL, Look MP, van Putten WLJ, et al. TP53 and MYC gene alterations independently predict poor prognosis in breast cancer patients. Genes Chromosomes Cancer 1996; 16: 170–179.
Berns EMJJ, van Staveren IL, Look MP, Smid M, Klijn JG, Foekens JA . Mutations in residues of TP53 that directly contact DNA predict poor outcome in human primary breast cancer. Br J Cancer 1998; 77: 1130–1136.
Sjogren S, Inganas M, Norberg T, Lindgren A, Nordgren H, Holmberg L, et al. The p53 gene in breast cancer: prognostic value of complementary DNA sequencing versus immunohistochemistry. J Natl Cancer Inst 1996; 88: 173–182.
Elledge RM, Allred DC . Prognostic and predictive value of p53 and p21 in breast cancer. Breast Cancer Res Treat 1998; 52: 79–98.
Pharoah PD, Day NE, Caldas C . Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br J Cancer 1999; 80: 1968–1973.
The uniform approach to breast fine-needle aspiration biopsy. National Cancer Institute Fine-Needle Aspiration of Breast Workshop Subcommittees. Diagn Cytopathol 1997;16:295–311.
Wilkinson EJ, Bland KI . Techniques and results of aspiration cytology for diagnosis of benign and malignant diseases of the breast. Surg Clin North Am 1990; 70: 801–813.
Bedard YC, Pollett AF . Breast fine-needle aspiration. A comparison of ThinPrep and conventional smears. Am J Clin Pathol 1999; 111: 523–527.
Leung SW, Bedard YC . Estrogen and progesterone receptor contents in ThinPrep-processed fine-needle aspirates of breast. Am J Clin Pathol 1999; 112: 50–56.
Leung SW, Bedard YC . Immunocytochemical staining on ThinPrep processed smears. Mod Pathol 1996; 9: 304–306.
Magda JL, Minger BA, Rimm DL . Polymerase chain reaction-based detection of clonality as a non-morphologic diagnostic tool for fine-needle aspiration of the breast. Cancer 1998; 84: 262–267.
Dimulescu I, Unger ER, Lee DR, Reeves WC, Vernon SD . Characterization of RNA in cytologic samples preserved in a methanol-based collection solution. Mol Diagn 1998; 3: 67–71.
Sherman ME, Schiffman MH, Lorincz AT, Herrero R, Hutchinson ML, Bratti C, et al. Cervical specimens collected in liquid buffer are suitable for both cytologic screening and ancillary human papillomavirus testing. Cancer 1997; 81: 89–97.
Elston CW, Ellis IO . Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 1991; 19: 403–410.
Zhu XL, Hartwick W, Rohan T, Kandel R . Cyclin D 1 gene amplification and protein expression in benign breast disease and breast carcinoma. Mod Pathol 1998; 11: 1082–1088.
Orita M, Suzuki Y, Sekiya T, Hayashi K . Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics 1989; 5: 874–921.
Murakami Y, Hayashi K, Sekiya T . Detection of aberrations of the p53 alleles and the gene transcript in human tumor cell lines by single-strand conformation polymorphism analysis. Cancer Res 1991; 51: 3356–3361.
Hainaut P, Hernandez T, Robinson A, Rodriguez-Tome P, Flores T, Hollstein M, et al. IARC Database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualisation tools. Nucleic Acids Res 1998; 26: 205–213.
Dawson-Saunders B, Trapp RG . Basic and clinical biostatistics. Norwalk, CT: Appleton and Lange; 1994.
Tiniakos DG, Robinson KB, Greenwood H, Cullen P, Cook AIM, Horne CHW, et al. c-erb B-2 and p53 expression in breast cancer fine needle aspirates. Cytopathology 1996; 7: 178–186.
Colecchia M, Frigo B, Zucchi A, Leopardi O . p53 protein expression in fine-needle aspirates of breast cancer: an immunocytochemical assay for identifying high-grade ductal carcinomas. Diagn Cytopathol 1995; 13: 128–133.
Makris A, Allred DC, Powles TJ, Dowsett M, Fernando IN, Trott PA, et al. Cytological evaluation of biological prognostic markers from primary breast carcinomas. Breast Cancer Res Treat 1997; 44: 65–74.
Williams C, Ponten F, Moberg C, Soderkvist P, Uhlen M, Ponten J, et al. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol 1999; 155: 1467–1471.
Wong C, DiCioccio RA, Allen HJ, Werness BA, Piver MS . Mutations in BRCA1 from fixed, paraffin-embedded tissue can be artifacts of preservation. Cancer Genet Cytogenet 1998; 107: 21–27.
Shiao YH, Buzard GS, Weghorst CM, Rice JM . DNA template as a source of artifact in the detection of p53 gene mutations using archived tissue. Biotechniques 1997;22:608–10,612.
Dunn JM, Hastrich DJ, Newcomb P, Webb JC, Maitland NJ, Farndon JR . Correlation between p53 mutations and antibody staining in breast carcinoma. Br J Surg 1993; 80: 1410–1412.
Saitoh S, Cunningham J, De Vries EM, McGovern RM, Schroeder JJ, Hartmann A, et al. p53 gene mutations in breast cancers in Midwestern US women: null as well as missense-type mutations are associated with poor prognosis. Oncogene 1994; 9: 2869–2875.
Osborne RJ, Merlo GR, Mitsudomi T, Venesio T, Liscia DS, Cappa AP, et al. Mutations in the p53 gene in primary human breast cancers. Cancer Res 1991; 51: 6194–6198.
Sato T, Yuyama Y, Watabe K, Okazaki A, Toda K, Okazaki M, et al. Detection of p53 gene mutations in fine-needle aspiration biopsied breast cancer specimens: correlations with nuclear p53 accumulation and tumor DNA aneuploidy patterns. Cancer Lett 1997; 115: 47–55.
Lavarino C, Corletto V, Mezzelani A, Della Torre G, Bartolli C, Riva C, et al. Detection of TP53 mutation, loss of heterozygosity and DNA content in fine-needle aspirates of breast carcinoma. Br J Cancer 1998; 77: 125–130.
Felley-Bosco E, Weston A, Cawley HM, Bennett WP, Harris CC . Functional studies of a germ-line polymorphism at codon 47 within the p53 gene. Am J Hum Genet 1993; 53: 752–759.
Sjalander A, Birgander R, Hallmans G, Cajander S, Lenner P, Athlin L, et al. p53 polymorphisms and haplotypes in breast cancer. Carcinogenesis 1996; 17: 1313–1316.
Carbone D, Chiba I, Mitsudomi T . Polymorphism at codon 213 within the p53 gene. Oncogene 1991; 6: 1691–1692.
Caleffi M, Teague MW, Jensen RA, Vnencak-Jones CL, Dupont WD, Parl FF . p53 gene mutations and steroid receptor status in breast cancer. Clinicopathologic correlations and prognostic assessment. Cancer 1994; 73: 2147–2156.
Soong R, Iacopetta BJ, Harvey JM, Sterrett GF, Dawkins HJ, Hahnel R, et al. Detection of p53 gene mutation by rapid PCR-SSCP and its association with poor survival in breast cancer. Int J Cancer 1997; 74: 642–647.
Pelosi G, Bresaola E, Rodella S, Manfrin E, Piubello Q, Schiavon I, et al. Expression of proliferating cell nuclear antigen, Ki-67 antigen, estrogen receptor protein, and tumor suppressor p53 gene in cytologic samples of breast cancer: an immunochemical study with clinical, pathobiological, and histologic correlations. Diagn Cytopathol 1994; 11: 131–140.
MacGeoch C, Barnes DM, Newton JA, Mohammed S, Hodgson SV, Ng M, et al. p53 protein detected by immunohistochemical staining is not always mutant. Dis Markers 1993; 11: 239–250.
Jacquemier J, Moles JP, Penault-Llorca F, Adelaide J, Torrente M, Viens P, et al. p53 immunohistochemical analysis in breast cancer with four monoclonal antibodies: comparison of staining and PCR-SSCP results. Br J Cancer 1994; 69: 846–852.
Acknowledgements
This work was supported in part by a grant from the US Army Medical Research and Materiel Command.
We thank Lori Cutler for her secretarial help.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Pollett, A., Bédard, Y., Li, SQ. et al. Correlation of p53 Mutations in ThinPrep-Processed Fine Needle Breast Aspirates with Surgically Resected Breast Cancers. Mod Pathol 13, 1173–1179 (2000). https://doi.org/10.1038/modpathol.3880216
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3880216