Abstract
Notch receptors regulate cell fate determination, stem cell self-renewal, proliferation and apoptosis. We previously reported that elevated mRNA expression of the Notch ligand JAG1 identifies breast cancer patients with a poor prognosis. Here we show through immunohistochemical analysis of the same breast cancer cases (N=127) that patients with tumors expressing high levels of JAG1 protein had a worse outcome than those with tumors expressing low levels (10-year survival 26 vs 48%, and median survival 63 vs 108 months, respectively; P=0.03). We also describe the novel application of the Allred score to quantify JAG1 mRNA and protein expression levels. Using the Allred score, patients with tumors expressing high levels of JAG1 mRNA had a worse outcome than those with tumors expressing low levels (10-year survival 16 vs 47%, and median survival 43 months vs 100 months, respectively; P<0.001). Interestingly, when tumors were classified as either high or low for JAG1 mRNA or protein expression, there was only 65% agreement (κ=0.08) between the two methods of expression analysis. When JAG1 mRNA and protein data were combined, patients with tumors expressing low levels of both had a 10-year survival of 53% and median survival of 131 months. In comparison, patients with tumors expressing either high levels of JAG1 protein, mRNA or both had reduced 10-year survival and median survival (31%, 19%, 11% and 77, 43, 23 months respectively; P<0.0001). There was marginal evidence of an interaction effect (P=0.055), which indicated that the prognostic value of JAG1 protein was limited to the JAG1 mRNA-low subgroup. These data show that the Allred score can be used to rapidly quantify JAG1 mRNA and protein levels in breast cancer to identify patients who have a significant survival disadvantage and who may benefit from therapies (such as γ-secretase inhibitors) that target signaling through the Notch pathway.
Similar content being viewed by others
Main
Breast cancer accounts for about 30% of malignancies diagnosed in North American women, skin cancer notwithstanding, it remains the second most common cause of cancer-related mortality.1, 2 Current treatment regimens are guided by outcome predictions based on clinical and pathological criteria established by the St Gallen, National Institutes of Health or Adjuvant! Online criteria.3, 4, 5 However, accumulating evidence suggests that breast cancer prognosis and response to therapy can be more accurately predicted by gene expression patterns characterizing specific breast cancer subtypes.6, 7, 8, 9, 10 We have recently identified a subtype of breast cancer characterized by elevated expression of mRNA encoding the Notch ligand JAG1, and poor patient outcome.11
The Notch signaling cascade is highly conserved and plays a crucial role in multiple cellular processes including stem cell self-renewal, cell fate determination, epithelial cell polarity/adhesion, cell division and apoptosis in organisms from worms to humans.12, 13, 14 Mammals have four Notch proteins (NOTCH1–4) that function as receptors for five Notch ligands (Dll1, 3, 4 and JAG1, 2). Binding of ligands to Notch leads to proteolytic cleavage of the receptor at a site just outside the plasma membrane by ADAM-family proteases. This is immediately followed by cleavage at a site just inside the plasma membrane. This second cleavage reaction by the presenilin-γ-secretase complex releases a cytoplasmic domain fragment from the plasma membrane, NOTCHIC (NIC).15, 16 NIC can then enter the nucleus and interact with the RBP-Jκ/CBF-1 transcription factor. This interaction converts CBF-1 from a transcription repressor to an activator, resulting in increased expression of genes including those which code for the HES and HEY family basic helix-loop-helix (bHLH) transcriptional regulatory proteins.13, 17, 18
Aberrant signaling within the Notch pathway is reported in multiple malignancies.19 The possibility that Notch may be important in human breast cancer comes from studies on mouse mammary tumor virus-induced neoplasms where Notch4 and Notch1 genes have been activated.20, 21, 22 In humans, loss of Numb, a NOTCH inhibitor, is thought to be important in a subset of breast tumors.23 In a separate study, the accumulation of NIC (and concomitant loss of Numb) was observed in a small group of lobular and ductal carcinomas of the breast, suggesting that Notch signaling was aberrantly activated in these tumors.24
We have demonstrated that JAG1 overexpression in carcinoma of the breast is associated with a poor prognosis.11 In that study we described a method using in situ hybridization and a complex image analysis algorithm to quantify JAG1 mRNA levels in paraffin-embedded breast cancer specimens. Here we report a simplified method to determine JAG1 levels through the application of the Allred score25, 26 to quantify JAG1 mRNA expression identified by in situ hybridization. In addition, we describe immunohistochemical analysis of JAG1 protein in breast cancer and demonstrate an association between elevated expression level and poor patient outcome. These studies support a role for mRNA and protein-based methods of JAG1 expression analysis in breast cancer prognostication.
Materials and methods
Breast Tumor Tissue
Breast cancer tissue microarray slides were obtained from the Cooperative Breast Cancer Tissue Resource (CBCTR), funded by the National Cancer Institute (NCI). The tissue microarrays included 192 samples of primary invasive breast cancer, with 64 cases each of node-negative, node-positive, and metastatic breast cancer. For 127 tumors, both JAG1 in situ hybridization and immunohistochemistry data were obtained (see below).
In Situ Hybridization
In situ hybridization analysis of breast cancer has previously been reported in detail elsewhere.11 Briefly, JAG1 cDNA probe for in situ hybridization was made using 33P-UTP-radiolabeled cRNA. Using routine techniques, the tissue microarray sections were hybridized with the radiolabeled antisense probe, washed and treated with Kodak NTB-2 nuclear emulsion. The tissue microarrays were stored at 4°C for several weeks prior to development in Kodak D-19 solution; they were subsequently fixed in Kodafix and counterstained with 0.1% toluidine blue.
Immunohistochemistry
Immunohistochemistry was performed using standard techniques. Briefly, 4-μm paraffin-embedded tissue microarray sections were dewaxed in xylene and rehydrated in graded alcohols. Endogenous peroxidase was blocked using 3% hydrogen peroxide. Antigen retrieval was accomplished using 10 mM citrate buffer (pH 6.0) in TT Mega Milestone (ESBE Scientific, Markham, Ontario, Canada). Nonspecific protein binding, avidin and biotin were blocked by 15 min incubations with goat serum, avidin-blocking reagent and biotin-blocking reagents (R&D, Minneapolis, MN, USA), respectively; these treatments were alternated with rinses in Tris-buffered saline (TBS). The slides were then treated with polyclonal goat anti-human JAG1 (1:10; R&D, Minneapolis, MN, USA) for 1 h at room temperature. Next, they were rinsed with TBS and incubated with biotinylated secondary antibody (R&D). This was followed by a rinse in TBS, incubation with HSS-HRP, DAB chromogen staining and counterstaining with Mayer's haematoxylin blue in Scott's water.
Quantification
mRNA expression of JAG1 was quantified in two ways: (i) image analysis as previously described11 and (ii) through application of the Allred score,25, 26 modified as follows: activated silver grains (resulting from hybridized 33P-UTP-radiolabeled cRNA) overlying and immediately adjacent to malignant cell nuclei were assigned an ‘intensity score’ (0=no activated silver grains; 1=weakly positive; 2=intermediately positive; 3=strongly positive). Next, the ‘proportion score’ was assigned for the dominant intensity pattern of in situ hybridization (0=no cells positive; 1=<1/100 cells positive; 2=1/100–1/10; 3=1/10–1/3; 4=1/3–2/3; 5=>2/3); combined, a ‘total score’ ranging from 0 to 8 was assigned to each tissue core.
Protein expression of JAG1 was quantified by immunohistochemical staining of the tissue microarray and application of the Allred score.
High-level expression was arbitrarily defined as JAG1 mRNA or protein expression in the upper quartile of the expression range. The top quartile of expression was determined by ranking the Allred scores for all tumors on the tissue microarray, and identifying the highest quarter. Since there were many tied scores, the top quartile of the expression range did not necessarily include a quarter of the tumors. For JAG1 mRNA, tumors with Allred scores ≥5 were in the top quartile of the expression range and were labeled ‘high’; for JAG1 protein expression, the top quartile of tumors had Allred scores ≥4.
Statistical Methods
In comparing the Allred Scoring method to that of image analysis, a κ measure of agreement was performed. The κ values ≤0.4 were considered to represent poor agreement, 0.4–0.75 good agreement and ≥0.75 excellent agreement. Overall survival was measured from diagnosis to last follow-up or death. Kaplan–Meier curves were calculated for the high and low expression of JAG1 mRNA, and protein. Survival between groups was compared using the log-rank test. The coexpression of high levels of JAG1 mRNA and protein were similarly investigated. Cox proportional hazard regression was used to look for a dose–response relationship between level of JAG1 expression and survival and to test whether JAG1 mRNA and protein level were independently related to survival. Hazard ratios (HRs) for continuous variables were expressed over a change equal to the size of the interquartile range (IQR; difference between the third quartile and first quartile values). P-values ≤0.05 were considered statistically significant.
Results
In Situ Hybridization and Application of the Allred Score to Quantify JAG1 mRNA Expression
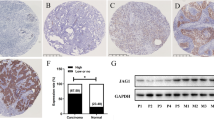
Our in situ hybridization expression analysis of JAG1 mRNA in breast cancers from a cohort of 192 patients on the CBCTR tissue microarray has been described elsewhere.11 In that study, the level of JAG1 mRNA expression for each tumor sample was determined from the concentration of activated silver grains overlying regions of malignant cells. This approach was based on a previous report demonstrating a linear relationship between the concentration of hybridized 33P-UTP-radiolabled cRNA probe and activated silver grain density.27 To quantify silver grain density in the breast cancers studied, we developed a highly reproducible but labor-intensive image analysis algorithm. Here we describe re-analysis of the same breast cancer specimens using the Allred method, modified to score silver grain density and distribution (Figure 1a and 1b).25, 26 This method was found to have good intra- and interobserver reliability (data not shown). By comparing these results to those previously obtained using image analysis, an 83% agreement for labeling tumors as expressing either high or low JAG1 mRNA levels was demonstrated (κ=0.51). Similarly, reanalysis of the CBCTR tissue microarray using a NOTCH1 cRNA probe demonstrated 82% agreement (κ=0.4) between image analysis and Allred quantitation.
Photomicrographs demonstrating the application of the Allred score to JAG1 expression. Tissue microarray cores that have undergone in situ hybridization with JAG1 cRNA probe (as identified by activated silver grains), Allred score 6 (a) and Allred score 0 (b). Tissue microarray cores showing immunohistochemistry for JAG1, Allred score 6 (c) and Allred score 0 (d). Photomicrographs are all shown using × 40 objective, with × 10 objective (inset).
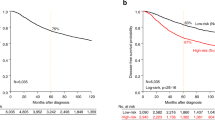
Similar to previous observations using image analysis, JAG1 expression level determined by the Allred method was found to correlate with a negative patient outcome. Patients whose tumors expressed high levels of JAG1 mRNA were found to have a significantly worse outcome compared to patients whose tumors expressed low levels of JAG1 (10-year survival 16 vs 47%, and median survival 43 vs 100 months, respectively; P<0.001) (Table 1; Figure 2a). Additionally, JAG1 expression analysis using the Allred score showed a continuous expression level-dependent relationship with negative outcome and this relationship was stronger than that found with image analysis (compare HR=2.18, 95% CI: 1.36–3.49 over IQR=4 units, P=0.0013 for Allred score vs HR=1.42, 95% CI: 1.03–1.95 over IQR=3 units, P=0.03 for image analysis). Furthermore, high JAG1 mRNA expression was found to be an independent predictor of poor outcome in bivariate analyses with other known predictors of outcome including patient age, metastases, tumor size, estrogen receptor positivity and tumor grade (data not shown). Bivariate analysis with lymph node status did not reach significance.
Kaplan–Meier curves showing relationship between high-level JAG1 mRNA and protein expression and overall survival in patients with breast cancer. Patients expressing high levels of JAG1 (high) mRNA scored using in situ hybridization and the Allred technique (a) or high levels of JAG1 protein scored using immunohistochemistry and the Allred technique (b), have a significantly shorter survival compared to those expressing low levels of JAG1 (low) (JAG1 in situ hybridization P<0.001; JAG1 immunohistochemistry P=0.03).
High-Level JAG1 Protein is Associated with Poor Outcome in Breast Cancer
In order to test whether JAG1 protein levels in breast cancer correlated with patient outcome, the CBCTR tissue microarray was screened using a commercially available JAG1 polyclonal antibody and expression quantified by applying the Allred score (Figure 1c and 1d). Analysis of the tumor tissues on the CBCTR tissue microarray revealed staining that was primarily cytoplasmic in nature; however, variable membranous and nuclear patterns were also observed. The scoring system was reproducible as an agreement of 82% (κ=0.50) was achieved when two independent reviewers scored the tissue microarrays. Outcome analysis revealed that patients with tumors expressing high levels of JAG1 protein had a worse outcome than those with tumors expressing low levels (10-year survival 26 vs 48%, and median survival 63 vs 108 months, respectively; P=0.03) (Table 1; Figure 2b). High JAG1 protein was found to be an independent predictor of poor outcome in bivariate analyses with age, estrogen receptor positivity and tumor grade. Bivariate analysis with tumor size reached borderline significance (P=0.06) and analyses with lymph node status and metastases did not reach significance for high JAG1 protein expression.
High-Level JAG1 mRNA and Protein Identify Two Distinct Subgroups of Patients with Poor-Outcome Breast Cancer
Having established that elevated expression of JAG1, analyzed either by mRNA in situ hybridization or by protein immunohistochemistry, predicted poor outcome in breast cancer, we wished to determine the measure of agreement between these two methods of expression analysis. While both tests identified 74 and nine (of 127 patients) as low and high for JAG1 expression, respectively (65% agreement), for 44 patients (35%) there was disagreement in assignment of expression level. An agreement measure of κ=0.08 indicated that the two methods of JAG1 expression analysis did not agree well and identified distinct subgroups of patients. We therefore compared outcome in the groups labeled as either high or low JAG1 expression by one or both tests. When JAG1 mRNA and protein data were combined, patients with tumors expressing low levels of both had a 10-year survival of 53% and median survival of 131 months (Table 1; Figure 3a). In contrast, compared to patients with low levels of both JAG1 mRNA and protein, patients with tumors expressing either high levels of JAG1 protein, mRNA or both had significantly reduced 10-year survival and median survival (31%, 19%, 11% and 77, 43, 23 months, respectively; P<0.0001) (Table 1; Figure 3a and b).
Relationship between combined JAG1 mRNA and protein levels and overall survival in patients with breast cancer. (a) Patients with tumors expressing low levels of JAG1 mRNA and protein had a 10-year survival of 53% and median survival of 131 months (—). (b) Patients with tumors expressing high levels of JAG1 mRNA and protein had a 10-year survival of 11% and median survival of 23 months (- - -). The value of adding protein data to predict outcome was exclusive to the JAG1 mRNA low group (interaction P=0.055).
We next examined whether high levels of JAG1 mRNA and protein were independent predictors of survival. In a bivariate Cox regression model, both JAG1 mRNA and protein remained significant (HR=2.5, 95% CI: 1.5–4.2, P<0.001 and HR=1.6, 95% CI: 1.0–2.6, P=0.05, respectively). There was marginal evidence of an interaction effect (P=0.055) which indicated that the prognostic value of JAG1 protein was limited to the JAG1 mRNA-low subgroup (Figure 3a and b). This is illustrated by the difference in the 10-year survival rates (Table 1). In the JAG1 mRNA low subgroup, the difference in 10-year survival between the low and high JAG1 protein groups was 22% (protein low: 53% vs protein high: 31%). In contrast, the survival difference in the JAG1 mRNA high subgroup was 8% (protein low: 19% vs protein high: 11%).
Discussion
Gene expression patterns are emerging as powerful tools for prognostication and predicting response to treatment in breast cancer. The present study confirms the utility of two methods to quantify JAG1 expression in paraffin-embedded breast cancer tissue for these purposes. The first method relies on in situ hybridization to determine JAG1 mRNA expression levels. The second is based upon immunohistochemical detection of JAG1 protein. Both methods involved the application of the Allred score to quantify JAG1 expression levels, and are readily applicable by clinical pathologists and investigators.
We demonstrated good agreement between our previously described image analysis technique and the Allred score to identify tumors having low vs high expression of JAG1 mRNA. Similarly, reanalysis of tissue microarrays probed with NOTCH1 cRNA using the Allred scoring system and originally scored by image analysis showed good agreement between the two methods. Our results demonstrate that use of the Allred score to interpret an in situ hybridization signal offers a rapid means to reproducibly quantify mRNA.
As was found previously using image analysis, Allred quantification of JAG1 in situ hybridization results on the CBCTR tissue microarray showed that patients whose tumors express high levels of JAG1 had a worse prognosis than those with low-level expression of JAG1 mRNA. In addition, the Allred score demonstrated a continuous expression level-dependent relationship with negative outcome that was stronger than that found with image analysis. The analysis of JAG1 protein levels on the CBCTR tissue microarray in this study reaffirmed the relationship between JAG1 expression and survival.
High JAG1 expression was found to be an independent predictor of poor outcome in bivariate analyses with all other known predictors of outcome tested (including patient age, estrogen receptor status, tumor size and tumor grade) with the exception of metastases (in the JAG1 protein analysis) and lymph node status (in both the JAG1 in situ hybridization and protein analyses). Sample size may have limited the predictive power of high JAG1 protein in the analysis with metastases, a marker of uniformly poor prognosis. Nodal status was missing in 25 of the 127 patients (19.7%) studied and the patients who had missing nodal status were disproportionate in the group with elevated JAG1 expression in both the in situ hybridization (32% of patients with missing nodal status had tumors with elevated JAG1 expression, while only 15% of patients not lacking nodal status had tumors with elevated JAG1 expression) and the immunohistochemical (40 and 28%, respectively) analyses. The relationship between missing nodal status and JAG1 level makes the observed lack of independence between the two variables less reliable.
The level of agreement between in situ hybridization and immunohistochemistry-based methods to identify tumors as high or low for JAG1 expression was poor. In a bivariate analysis with JAG1 mRNA expression, JAG1 protein level was found to be an independent predictor of outcome. These data suggest that elevated JAG1 mRNA and protein expression identify overlapping, but distinct subgroups of patients with poor-outcome breast cancer.
There are several possibilities to explain the disagreement between in situ hybridization and immunohistochemistry-based methods of JAG1 quantification. Biologic explanations for the disagreement may include tumor-specific differences in JAG1 mRNA/protein stability or differences in post-transcriptional regulation of JAG1 expression. Technical causes contributing to the disagreement may relate to sample fixation,28 age of stored samples,28 nature of antibody,29 specificity of the cRNA probe or antigen retrieval technique.30 Heterogeneous concordance rates between mRNA and protein expression, for various reasons, have been reported in numerous systems.31, 32, 33, 34, 35 In adenocarcinoma, variable rates of concordance have been observed when comparing qualitative real-time RT-PCR, immunohistochemistry and FISH-based methods of HER2/neu expression analysis.30, 36, 37 Without further studies, it remains difficult to explain the present disagreement between in situ hybridization and immunohistochemistry-based methods of JAG1 quantification.
While the reason for the discordance between in situ hybridization and immunohistochemistry-based methods of JAG1 quantification is unexplained, it is clear that patients with tumors expressing high levels of JAG1 (mRNA, protein or both) have worse outcome than patients whose tumors show no evidence of JAG1 overexpression. In this context, it is interesting that we identified nine patients with tumors demonstrating combined high-level JAG1 mRNA and protein and who demonstrated the worst 10-year and median survival of all subgroups in our study (11% and 23 months, respectively). The present study was not sufficiently powered to determine whether this finding is statistically significant but this trend will be further explored in future work.
In summary, we have confirmed that JAG1 mRNA and protein are useful biomarkers in breast cancer, and indicative of a poor patient outcome; this, using a simplified method of quantification based on the Allred score. This technique is reproducible, efficient and readily applicable in a research or clinical setting. JAG1 overexpression is increasingly recognized in human cancers and may have a role as a prognostic marker or as a predictor of chemotherapy response.38 JAG1 in situ hybridization and/or immunohistochemistry may be useful alone or in combination with other known biomarkers as part of a prognostic/predictive biomarker platform. Furthermore, high-level JAG1 expression may identify tumors susceptible to treatments that target the Notch pathway. Potential approaches may include: γ-secretase inhibitors, which exploit the dependence of NOTCH receptor on enzyme processing for its activity; Notch ligand/Fc fusion proteins that inhibit binding and signaling through the NOTCH receptor, and inhibitory siRNA structures that target mRNA encoding NOTCH receptors or their ligands.39
References
American Cancer Society. Cancer facts and figures 2006. Atlanta, 2006. http://www.cancer.org/downloads/STT/CAFF2006PWSecured.pdf.
Canadian Cancer Society/National Cancer Institute of Canada. Canadian Cancer Statistics 2006. Toronto, 2006. http://www.cancer.ca/vgn/images/portal/cit_86751114/31/21/935505792cw_2006stats_en.pdf.pdf.
Goldhirsch A, Glick JH, Gelber RD, et al. Meeting highlights: International Consensus Panel on the Treatment of Primary Breast Cancer. Seventh International Conference on Adjuvant Therapy of Primary Breast Cancer. J Clin Oncol 2001;19:3817–3827.
Eifel P, Axelson JA, Costa J, et al. National Institutes of Health Consensus Development Conference Statement: adjuvant therapy for breast cancer, November 1–3, 2000. J Natl Cancer Inst 2001;93:979–989.
Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001;98:10869–10874.
Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res 2005;11:5678–5685.
Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817–2826.
van't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002;415:530–536.
Chang HY, Nuyten DS, Sneddon JB, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci USA 2005;102:3738–3743.
Reedijk M, Odorcic S, Chang L, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res 2005;65:8530–8537.
Artavanis-Tsakonas S, Matsuno K, Fortini ME . Notch signaling. Science 1995;268:225–232.
Artavanis-Tsakonas S, Rand MD, Lake RJ . Notch signaling: cell fate control and signal integration in development. Science 1999;284:770–776.
Egan SE, St-Pierre B, Leow CC . Notch receptors, partners and regulators: from conserved domains to powerful functions. Curr Top Microbiol Immunol 1998;228:273–324.
Iwatsubo T . The gamma-secretase complex: machinery for intramembrane proteolysis. Curr Opin Neurobiol 2004;14:379–383.
Xia W, Wolfe MS . Intramembrane proteolysis by presenilin and presenilin-like proteases. J Cell Sci 2003;116:2839–2844.
Iso T, Kedes L, Hamamori Y . HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol 2003;194:237–255.
Maier MM, Gessler M . Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem Biophys Res Commun 2000;275:652–660.
Weng AP, Aster JC . Multiple niches for Notch in cancer: context is everything. Curr Opin Genet Dev 2004;14:48–54.
Jhappan C, Gallahan D, Stahle C, et al. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev 1992;6:345–355.
Gallahan D, Jhappan C, Robinson G, et al. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res 1996;56:1775–1785.
Dievart A, Beaulieu N, Jolicoeur P . Involvement of Notch1 in the development of mouse mammary tumors. Oncogene 1999;18:5973–5981.
Pece S, Serresi M, Santolini E, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol 2004;167:215–221.
Stylianou S, Clarke RB, Brennan K . Aberrant activation of notch signaling in human breast cancer. Cancer Res 2006;66:1517–1525.
Allred DC, Clark GM, Elledge R, et al. Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J Natl Cancer Inst 1993;85:200–206.
Harvey JM, Clark GM, Osborne CK, et al. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol 1999;17:1474–1481.
Swan MC, Najlerahim AR, Bennett JP . Expression of serotonin transporter mRNA in rat brain: presence in neuronal and non-neuronal cells and effect of paroxetine. J Chem Neuroanat 1997;13:71–76.
Atkins D, Reiffen KA, Tegtmeier CL, et al. Immunohistochemical detection of EGFR in paraffin-embedded tumor tissues: variation in staining intensity due to choice of fixative and storage time of tissue sections. J Histochem Cytochem 2004;52:893–901.
Press MF, Hung G, Godolphin W, et al. Sensitivity of HER-2/neu antibodies in archival tissue samples: potential source of error in immunohistochemical studies of oncogene expression. Cancer Res 1994;54:2771–2777.
Saxby AJ, Nielsen A, Scarlett CJ, et al. Assessment of HER-2 status in pancreatic adenocarcinoma: correlation of immunohistochemistry, quantitative real-time RT-PCR, and FISH with aneuploidy and survival. Am J Surg Pathol 2005;29:1125–1134.
Anderson L, Seilhamer J . A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 1997;18:533–537.
Lichtinghagen R, Musholt PB, Lein M, et al. Different mRNA and protein expression of matrix metalloproteinases 2 and 9 and tissue inhibitor of metalloproteinases 1 in benign and malignant prostate tissue. Eur Urol 2002;42:398–406.
Chen G, Gharib TG, Huang CC, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics 2002;1:304–313.
Orntoft TF, Thykjaer T, Waldman FM, et al. Genome-wide study of gene copy numbers, transcripts, and protein levels in pairs of non-invasive and invasive human transitional cell carcinomas. Mol Cell Proteomics 2002;1:37–45.
Greenbaum D, Colangelo C, Williams K, et al. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol 2003;4:117.
Perez EA, Roche PC, Jenkins RB, et al. HER2 testing in patients with breast cancer: poor correlation between weak positivity by immunohistochemistry and gene amplification by fluorescence in situ hybridization. Mayo Clin Proc 2002;77:148–154.
Dal Lago L, Durbecq V, Desmedt C, et al. Correction for chromosome-17 is critical for the determination of true Her-2/neu gene amplification status in breast cancer. Mol Cancer Ther 2006;5:2572–2579.
Zhang Y, Wang Z, Ahmed F, et al. Down-regulation of Jagged-1 induces cell growth inhibition and S phase arrest in prostate cancer cells. Int J Cancer 2006;119:2071–2077.
Purow BW, Haque RM, Noel MW, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res 2005;65:2353–2363.
Acknowledgements
MR was supported by a Junior Faculty Grant Award from the Society of University Surgeons. SE was supported by a grant from the National Cancer Institute of Canada with funds from the Terry Fox Run. The authors thank Suzanna Tjan for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors report no conflicts of interest.
Rights and permissions
About this article
Cite this article
Dickson, B., Mulligan, A., Zhang, H. et al. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol 20, 685–693 (2007). https://doi.org/10.1038/modpathol.3800785
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800785
Keywords
This article is cited by
-
Jagged1 intracellular domain/SMAD3 complex transcriptionally regulates TWIST1 to drive glioma invasion
Cell Death & Disease (2023)
-
Estrogen-dependent DLL1-mediated Notch signaling promotes luminal breast cancer
Oncogene (2019)
-
Role of the Nervous System in Tumor Angiogenesis
Cancer Microenvironment (2018)
-
Albumin uptake in human podocytes: a possible role for the cubilin-amnionless (CUBAM) complex
Scientific Reports (2017)
-
Evidence of novel miR-34a-based therapeutic approaches for multiple myeloma treatment
Scientific Reports (2017)