Abstract
Astrocytic gliomas are the most common pediatric brain tumors; however, nonbrainstem glioblastomas are extremely rare compared with their adult counterparts. Little information is available on the clinical significance of various molecular markers in pediatric grade IV astrocytomas. The current study was focused on the molecular analysis and clinico-pathological correlations in a set of 44 tumor samples obtained from pediatric patients with nonbrainstem glioblastomas. Fluorescence in situ hybridization (FISH) with a set of 10 commercial chromosome probes (1p36, 1q25, centromere (CEP)7, EGFR, CEP9, 9p21/p16, CEP10, 10q23/PTEN, 19p13, and 19q13) was performed. Disclosed molecular abnormalities, in descending order of frequency, included polysomy 7 (72%), loss of 10q23 (61%), loss of 9p21 (52%), loss of 1p36 (41%), gain of 1q25 (25%), polysomy 9 (16%), EGFR amplification (9%), loss of 19q13 (5%), polysomy 19 (5%), and codeletion 1p36/19q13 (2%). The overall survival time was markedly shorter only for those patients whose lesions harbored deletion of 10q23/PTEN locus (log-rank test; P=0.00007). By multivariate analysis, only loss of 10q23 locus reached an independent level of prognostic value (hazard ratio=2.88; P=0.01). There were no significant differences in patient survival for other molecular abnormalities. In conclusion, a FISH analysis of 10q23 dosage should be recommended as an ancillary laboratory method that allows further clinical subdivision of pediatric glioblastomas.
Similar content being viewed by others
Main
Astrocytic gliomas are the most common pediatric brain tumors; however, nonbrainstem glioblastomas are extremely rare compared with their adult counterparts and consist of approximately 3% of all neoplasms of the central nervous system in childhood. The outcome for children with supratentorial glioblastomas remains poor even with current therapeutic protocols and the prognostic value of various clinical parameters has been analyzed for these tumors. Thus far, the only consistent prognostic variable includes the total tumor removal independent of tumor location, patient age, and adjuvant treatment.1, 2, 3
In order to optimize treatment strategies for pediatric glioblastoma patients, a more precise understanding of the cellular and molecular basis of this disease is clearly necessary. Over the past several years, a variety of numerical chromosomal abnormalities have been described in adult glioblastomas, including gain of chromosome 7, and deletions involving chromosomes 1, 9, 10 and 19.3 Additionally, amplifications of various potent oncogenes (EGFR, CDK4, MDM2) and deletions/mutations of putative tumor suppressor genes (TP53, PTEN, CDKN2A/p16) have been implicated in adult glioblastoma molecular biology.3 Nevertheless, relatively few studies have been performed on molecular properties of pediatric glioblastomas, while different genetic pathways have been suggested in the tumorigenesis of childhood and adult malignant gliomas.4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15 Also, little information is available on the clinical significance of various molecular markers in pediatric high-grade astrocytomas. A few retrospective studies have found associations between adverse outcomes and p53 alterations,8 gain of 1q,13 and mutation of PTEN gene-suppressor.11 In contrast to adult malignant gliomas, deletions involving chromosomal loci 1p and 19q did not predict clinical outcome in their pediatric counterparts.10 The various histological subtypes of high-grade gliomas (anaplastic astrocytomas, glioblastomas, and mixed gliomas) were simultaneously analyzed in all studies mentioned above.
Our current study was focused on the molecular analysis and clinico-pathological correlations in a set of 44 tumor samples obtained from patients with nonbrainstem glioblastomas whose age at the time of diagnosis was less than 18 years. Interphase fluorescence in situ hybridization (FISH) was utilized for this purpose using a set of 10 commercially available chromosome probes (both centromere and locus-specific).
Materials and methods
Patient Population and Pathological Analysis
A total of 44 patients younger than 18 years who were treated in the Burdenko Neurosurgical Institute from January 1, 1994, to January 1, 2004, and who had newly diagnosed, supratentorial glioblastomas were included in this study. According to the current histological WHO classification,3 these were high-grade astrocytic gliomas showing brisk mitotic activity, prominent microvascular proliferation, and necrosis with pseudopalisades. In all cases, immunohistochemistry with the antibody to glial fibrillary acidic protein ((GFAP) (clone 6F2, prediluted, DakoCytomation, Carpinteria, CA, USA) was performed to confirm the astrocytic nature of the lesions. For differential diagnosis with primitive neuroectodermal tumors, the antibodies to synaptophysin (clone SY-38, 1:200, DakoCytomation), neurofilament proteins (clone 2F11, prediluted, DakoCytomation), and beta tubulin (clone D10, 1:100, Santa Cruz Biotechnology, Santa Cruz, CA, USA) were also applied.
The end point of the follow-up analysis was January 1, 2004, and the dates of death or last contact were considered the end of the study. Data on overall survival are shown as both the median survival times and the 5-year survival rates. The Institutional Review Board obtained approval to link laboratory data to clinical data.
Fluorescence In Situ Hybridization
A two-color FISH assay was performed on 5-μm-thick sections. The following four commercial sets of fluorochrome-labelled probes were applied (all produced by Vysis, Inc., Abbott Laboratories SA, Downers Grove, IL, USA: centromere (CEP)7/EGFR dual color probe set (Cat. No. 32-191053), CEP9/9p21 dual color probe set (Cat. No. 32-190078), CEP10/10q23 dual color probe set (Cat. No. 32-231010), and mixed 1p36/1q25 and 19p13/19q13 dual color probe sets (Cat. No. 32-231004). After protease pretreatment of the slides and its fixation, the probes (10 μl per slide) were applied to the sections. Simultaneous probe/specimen denaturation at 76°C for 8 min with subsequent overnight incubation at 37°C was performed in the HYBriteTM hybridization chamber (Vysis Inc.). Posthybridization process included subsequent washing in 50% formamide/2 × SSC (5 min at 42°C × 4) and 0.5 × SSC with NP40 (5 min at 60°C × 3). Nuclei were counterstained with DAPI (1000 ng DAPI/ml in antifade mounting solution, Vysis Inc.).
The sections were studied with an Axioplan 2 fluorescent microscope (Karl Zeiss, Gottingen, Germany) that was equipped with a set of the appropriate filters (Vysis Inc.). Signal detection was performed as described previously.16, 17, 18, 19, 20 Briefly, samples showing sufficient FISH efficiency (>90% nuclei with signals) were evaluated. Two independent referees (AK and RS) were unaware of clinical data. Signals were scored in at least 200 nonoverlapping, intact nuclei. Non-neoplastic temporal lobe specimens (n=8) were used as a control for each probe pair. Specimens were considered amplified for EGFR when more than 10% of tumor cells exhibited either EGFR:CEP7 ratio >2 or innumerable tight clusters of signals of the locus probe. Hemizygous deletions were defined as >50% nuclei (mean±3 standard deviations in nonneoplastic controls) containing either one signal of locus-targeted probe and ≥2 signals of reference probe (absolute deletions) or two signals of locus-targeted probe and ≥4 signals of reference probe (relative deletions). Homozygous deletions were identified by the simultaneous lack of both signals of locus-targeted probe and the presence of signals reference probe in more than 30% of cells. Monosomy for chromosomes 9 and 10 was defined by the presence of one CEP signal per cell in >50% (mean±3 standard deviations in non-neoplastic controls). Polysomies (chromosomal gains) were defined as >10% of nuclei containing three or more signals for locus or CEP, as no such findings were found in the control brain tissue.
Images were captured by using a high-resolution CCD microscopy camera AxioCam MRm REV2 (Karl Zeiss). The resulting images were reconstructed with green (FITC), red (Cy3), and blue (DAPI) pseudocolors using AxioVision 4 multichannel fluorescence basic workstation (Karl Zeiss) according to the manufacturer's instructions.
According to prior experience17, 18 homozygous and hemizygous deletions, as well as absolute and relative hemizygous deletions, were considered biologically equivalent.
Statistical Analysis
Nonparametric Spearman's rank correlation coefficients were used to assess the degree of linear associations between pairs of molecular markers. Survival analyses from the date of operation were estimated with the Kaplan–Meier method. The comparisons among various patient subgroups were performed by the log-rank test. Multivariate analysis for survival was performed using the Cox proportional hazard models. A significant correlation between two parameters was taken at the 95% confidence interval. Probability (P) values <0.05 were considered significant.
Results
Clinical Variables and Follow-Up Data
The 44 patients included 24 males and 20 females. The age of patients ranged from 2 to 17 years with a median age of 12.3 years, and 16 patients (37%) were younger than 10 years. According to preoperative contrast computed tomography and/or magnetic resonance imaging data, the tumor occupied the temporal lobe in 19 cases, the parietal lobe in 11 cases, the basal ganglia in seven cases, the frontal lobe in six cases, and the occipital lobe in the remaining one patient. In all the cases, an intense and heterogeneous ring-like contrast enhancement was identified. All of our patients had undergone open surgery with either macroscopically total/subtotal (32 cases) or partial (12 cases) tumor resection that was confirmed by postoperative neuroradiological studies. Postoperative Karnofsky performance scores were more than 70. The 42 patients older than 3 years received postoperative radiotherapy (external beam irradiation) with a total dose 45–52 cGy (mean 48 cGy) and all 44 patients received chemotherapy. At the date of end point of the follow-up, 29 patients (66%) had died within the period of 3–34 months after surgery; the median survival time was 11.4 months. Survival time for the remaining 14 survivors varied from 8 to 98 months with a median survival time of 23.4 months. The median overall survival for the entire cohort of patients was 15.7 months and the 5-year survival rate was 18%. By univariate analysis, we were unable to find any significant association between the patient overall survival and clinical variables including age less than or greater than 10 years, gender, tumor location, and volume of resection.
Data of FISH Analysis and Clinico-Pathological Associations
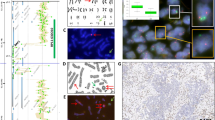
Representative results of FISH analysis are presented in Figure 1. Of the 210 total paired hybridizations, all yielded interpretable results. The most frequently encountered alteration was polysomic gain of chromosome 7, which was found in 32 cases (72%). Other frequent molecular abnormalities, in descending order of frequency, included deletion of 10q23/PTEN (27 samples; 61%) accompanied by monosomy 10 in nine cases, deletion of 9p21/p16 (23 samples; 52%) accompanied by monosomy 9 in two cases, and isolated deletion of 1p36 (18 samples; 41%). Most often there were absolute deletions. The vast majority (78%) of 9p21 deletions were homozygous, whereas no homozygous deletions of 10q23 were found. Other detectable alterations were gain of 1q25 (11 samples; 25%), polysomy 9 (seven samples; 16%), EGFR amplification (four samples; 9%) isolated deletion of 19q13 (two samples, 5%), gain of entire chromosome 19 (two samples, 5%), and combined loss of 1p36 and 19q13 (one sample; 2%). None of the tumors examined disclosed balanced (diploid) cytogenetic profile for chromosomes 1, 7, 9, 10, and 19 simultaneously. Correlation analysis revealed a significant association only between gain of 7 and loss of 10q (r=0.63; P<0.001).
Representative appearance of the frequent molecular abnormalities detected by FISH in pediatric glioblastomas. (a) Hemizygous deletion of 10q23/PTEN locus—single red signals in most of nuclei and paired green signals for CEP10. (b) Homozygous deletion of 9p21/p16 locus—absence of red signals in most of the nuclei and retained green signals for CEP9. (b) Deletion of 1p36 (single red signals in most of the nuclei) with simultaneous gain of 1q25 (green signals). (d) EGFR amplification (red signals and clouds) and polysomy 7 (green signals).
After acquisition of FISH data, the cases examined were reviewed to search for potential associations between clinical variables and discerned molecular features. As a result, we found no significant associations between molecular findings and patient age (less than or greater than 10 years), gender, and tumor location.
The survival time was markedly shorter only for those patients whose lesions harbored deletion of 10q23/PTEN locus (log-rank test; P=0.00007; Figure 2). In addition, by multivariate analysis, only loss of 10q23 locus reached an independent level of prognostic value (hazard ratio=2.88; P=0.01). There were no significant differences in patient survival for other molecular abnormalities. Also, the survival times did not differ significantly for tumors with isolated 10q23/PTEN loss and those that showed monosomy 10.
Discussion
Childhood glioblastomas are very rare in comparison with their adult counterparts, and the prognosis of these neoplasms still remains a controversial issue. Accordingly, these tumors consisted of the small minority (3.2%) of all glioblastomas diagnosed at our department over the last 10 years. The current study showed that the 5-year survival rate of 18% calculated for the cohort of 44 pediatric glioblastoma patients is greater than those reported previously for the entire populations of these malignancies.3, 21, 22 Approximately 70% of the patients from the current series died rapidly despite aggressive treatment with multimodal therapy, but a few other patients revealed a stable course of the disease with a relatively long overall survival. Thus, histologically uniform nonbrainstem glioblastomas of childhood vary greatly in terms of their biologic behavior and we hypothesized that the further clinical subdivision of these tumors could be achieved more precisely on the basis of molecular analysis.
A few previous reports have shown that FISH is a rapid and sensitive method of genetically stratifying astrocytic gliomas that provides prognostically useful information.16, 17, 18, 19, 20 In the current study, we applied FISH technique for molecular profiling of pediatric glioblastomas, keeping in the mind its applicability to paraffin-embedded specimens and the presence of well-established criteria for reproducible results interpretation. To the best of our knowledge, this is a first FISH analysis of a representative cohort of pediatric glioblastomas. The careful patient selection including similar treatment regimens and postoperative performance scores ensured that evaluation of the significance of molecular findings in predicting the clinical outcome was likely to be objective and reliable.
We found that the frequencies of polysomy 7, loss of 9p21/p16, and loss of 10q23/PTEN in pediatric glioblastomas correlate well with those that have been previously identified in their adult counterparts using FISH as well as various other molecular diagnostic techniques.18, 22, 23, 24, 25, 26 Nevertheless, the frequencies of some other chromosomal alterations varied strongly between adult and pediatric glioblastomas, supporting the notion that these histologically similar tumor entities can develop and progress along substantially different molecular pathways.
The high frequency of various alterations involving chromosome 1 has been already noted in pediatric glioblastomas.10, 13 Rickert et al.13 studied 13 specimens using of comparative genome hybridization and found that gain of 1q was the most frequent chromosomal aberration affecting 54% of these tumors. In contrast, we revealed this chromosomal alteration in only 25% of pediatric glioblastomas and this discrepancy may be explained by differences in the experimental design and number of tumors examined. Moreover, approximately 40% of pediatric glioblastomas from the current series revealed loss of 1p36 locus, although the frequency of such alteration in adult high-grade astrocytic gliomas does not exceed 15%.16, 17, 18, 19, 22, 23, 24, 25, 26 These findings allow one to suggest the presence of tumor suppressor gene(s) located on 1p36, an alteration that may be involved in carcinogenesis of pediatric glioblastomas, as it has been identified in embryonal neuroepithelial tumors of childhood.3
Chromosome 19 is one of the most commonly altered chromosomes in glioblastomas, while gain of 19 was previously found closely related with older patients and adverse outcomes.3, 24, 26 In contrast, loss of 19q has been established to be a hallmark of glioblastomas that may develop through a secondary multistep transformation process, and this abnormality was associated with long-term survival.3, 16, 24, 26 Nevertheless, various aberrations of chromosome 19 were found in the small minority of tumors from our current series, suggesting that these molecular alterations play no significant role in the evolution of pediatric glioblastomas, as well as codeletion of 1p/19q loci that was identified only in one tumor specimen.
It has been postulated previously that EGFR amplification is much less common in pediatric glioblastomas than in these tumors in adults, although only a few studies have reported data from analysis of this oncogene amplification in high-grade gliomas of childhood. In some of them, no EGFR amplifications were observed among pediatric glioblastomas,5, 11, 13, 14 whereas in a few small series, the frequency of this aberration reached 25%.6, 12 Finally, Bredel et al 4 have found EGFR amplification in two of 27 supratentorial pediatric glioblastomas (7.4%), which is a rate quite compatible with that observed by us in the current series (9%). Owing to the small number of cases, no clear association between EGFR amplification and patient outcome was apparent.
There are conflicting results regarding the prognostic value of various molecular alterations in the subject of adult glioblastomas.3, 16, 22, 23, 24, 25, 26 As has been pointed out earlier, only a few studies have evaluated the clinical significance of various molecular biologic patterns for pediatric high-grade gliomas.4, 7, 8, 10, 11, 13 In the present study, only loss of 10q23/PTEN locus was found significantly and independently associated with adverse outcome of childhood glioblastomas, while a few recent reports showed deletion of 10q as a negative prognostic indicator for their adult counterparts.22, 24, 25, 26 In addition, our findings are in line with data obtained by Raffel et al.11 These investigators found that mutation of PTEN was a single genetic alteration that revealed an association with survival of childhood with malignant gliomas, whereas other molecular abnormalities (TP53 mutation, CDKN2A deletion, CDK4 amplification) had no prognostic value.
In summary, despite the various genetic alterations existing in pediatric glioblastomas, only a FISH analysis of 10q23 dosage should be recommended after independent confirmation by other studies as an ancillary laboratory method that allows further clinical subdivision of these tumors. It gives evidence that the panel of molecular markers applied up to date is insufficient to cover an entire spectrum of the clinically significant genetic alterations in glioblastomas of childhood. Perhaps microarray-based screening of these infrequent brain malignancies will be necessary to identify the candidate genes and chromosomal regions having both prognostic and therapeutic relevance.
References
Broniscer A, Gajjar A . Supratentorial high-grade astrocytoma and diffuse brainstem glioma: two challenges for pediatric oncologist. Oncologist 2004;9:197–206.
Reddy AT, Wellons III JC . Pediatric high-grade gliomas. Cancer J 2003;9:107–112.
Kleihues P, Cavenee WK . Tumors of the Nervous System. Pathology and Genetics: World Health Organization International Classification of Tumours. WHO/IARC: Lyon, France, 2000.
Bredel M, Pollack IF, Hamilton RL, et al. Epidermal growth factor receptor expression and gene amplification in high-grade non-brain stem gliomas of childhood. Clin Cancer Res 1999;5:1786–1792.
Cheng Y, Ng HK, Zhang SF, et al. Genetic alterations in pediatric high-grade astrocytomas. Hum Pathol 1999;30:1284–1290.
Di Sapio A, Morra I, Pradotto L, et al. Molecular genetic changes in a series of neuroepithelial tumors of childhood. J Neuro-Oncol 2002;59:117–122.
Gilbertson RJ, Hill DA, Hernan R, et al. ERBB1 is amplified and overexpressed in high-grade diffusely infiltrative pediatric brain stem glioma. Clin Cancer Res 2003;9:3620–3624.
Pollack IF, Finkelstein SD, Woods J, et al. Expression of p53 and prognosis in children with malignant gliomas. N Engl J Med 2002;346:420–427.
Pollack IF, Finkelstein SD, Burnham JM, et al. Age and TP53 mutation frequency in childhood malignant gliomas: results in a multi-institutional cohort. Cancer Res 2001;61:7404–7407.
Pollack IF, Finkelstein SD, Burnham JM, et al. Association between chromosome 1p and 19q loss and outcome in pediatric malignant gliomas: results form the CCG-945 cohort. Pediatr Neurosurg 2003;39:114–121.
Raffel C, Frederick L, O'Fallon JR, et al. Analysis of oncogene and tumor suppressor gene alterations in pediatric malignant astrocytomas reveals reduced survival for patients with PTEN mutations. Clin Cancer Res 1999;5:4085–4090.
Rasheed BK, McLendon RE, Herndon JE, et al. Alterations of the TP53 gene in human gliomas. Cancer Res 1994;54:1324–1330.
Rickert CH, Strater R, Kaatsch P, et al. Pediatric high-grade astrocytomas show chromosomal imbalances distinct from adult cases. Am J Pathol 2001;158:1525–1532.
Sung T, Miller DC, Hayes RL, et al. Preferential inactivation of the p53 tumor suppressor pathway and lack of EGFR amplification distinguish de novo high-grade pediatric astrocytomas from de novo adult astrocytomas. Brain Pathol 2000;10:249–259.
Sure U, Ruedi D, Tachibana O, et al. Determination of p53 mutation, EGFR overexpression, and loss of p16 expression in pediatric glioblastomas. J Neuropathol Exp Neurol 1997;56:782–789.
Brat DJ, Seiferheld WF, Perry A, et al. Analysis of 1p, 19q, 9p and 10q as prognostic markers for high-grade astrocytomas using fluorescence in situ hybridization on tissue microarrays from Radiation Therapy Oncology Group. Neuro-Oncology 2004;6:96–103.
Fuller CE, Schmidt RE, Roth KA, et al. Clinical utility of fluorescence in situ hybridization (FISH) in morphologically ambiguous gliomas with hybrid oligodendroglial/ astrocytic features. J Neuropathol Exp Neurol 2003;62:1118–1128.
Fuller CE, Wang H, Zhang W, et al. High-throughput molecular profiling of high-grade astricytomas: the utility of fluorescence in situ hybridization on tissue microarrays (TMA-FISH). J Neuropathol Exp Neurol 2002;61:1078–1084.
Korshunov A, Golanov A, Sycheva R . Molecular stratification of diagnostically challenging high-grade gliomas composed of small cells. The utility of fluorescence in situ hybridization. Clin Cancer Res 2004;10:7820–7826.
Perry A, Aldape KD, George DH, et al. Small cell astrocytoma: an aggressive variant that is clinicopathologically and genetically distinct from anaplastic oligodendroglioma. Cancer 2004;100:2318–2326.
Lamborn KR, Chang SM, Prados MD . Prognostic factors for survival of patients with glioblastoma. Recursive portioning analysis. Neuro-Oncology 2004;6:227–235.
Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res 2004;64:6892–6899.
Batchelor TT, Betensky RA, Esposito JM, et al. Age-dependent prognostic effects of genetic alterations in glioblastomas. Clin Cancer Res 2004;10:228–233.
Burton E, Lamborn KR, Feuerstein BG, et al. Genetic aberrations defined by comparative genome hybridization distinguish long-term from typical survivors in glioblastoma. Cancer Res 2002;62:6205–6210.
Schmidt MC, Antweiler S, Urban N, et al. Impact of genotype and morphology on the prognosis of glioblastoma. J Neuropathol Exp Neurol 2002;61:321–328.
Wiltshire RN, Herndon JE, Lloyd A, et al. Comparative genomic hybridization analysis of astrocytomas. Prognostic and diagnostic implications. J Mol Diagn 2004;6:166–179.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Korshunov, A., Sycheva, R., Gorelyshev, S. et al. Clinical utility of fluorescence in situ hybridization (FISH) in nonbrainstem glioblastomas of childhood. Mod Pathol 18, 1258–1263 (2005). https://doi.org/10.1038/modpathol.3800415
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800415
Keywords
This article is cited by
-
Association of ERG/PTEN status with biochemical recurrence after radical prostatectomy for clinically localized prostate cancer
Medical Oncology (2018)
-
Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers
Acta Neuropathologica (2015)
-
SOX4 is associated with poor prognosis in prostate cancer and promotes epithelial–mesenchymal transition in vitro
Prostate Cancer and Prostatic Diseases (2013)
-
PTEN losses exhibit heterogeneity in multifocal prostatic adenocarcinoma and are associated with higher Gleason grade
Modern Pathology (2013)
-
Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression
Modern Pathology (2009)