Abstract
Activating point mutations of the BRAF oncogene have been identified in several solid tumors, most commonly in cutaneous melanomas and papillary carcinomas of the thyroid. A specific point mutation—V599E—accounts for the overwhelming majority of these mutational events. We explored the frequency of the V599E BRAF mutation in biliary tract cancers. In all, 62 archival biliary tract cancers, including 15 gallbladder cancers, 15 extrahepatic, and 10 intrahepatic cholangiocarcinomas from the United States, and 22 gallbladder carcinomas from Chile were analyzed for the V599E mutation of the BRAF gene using three distinct methods: direct sequencing, a primer extension method (Mutector® assay), and the highly sensitive quantitative Gap Ligase Chain Reaction. The common V599E mutation was not identified in any of the 62 biliary cancer samples using these three methods of detection. The V599E somatic mutation of the BRAF gene is absent in biliary tract cancers, at least in the two geographic populations (United States and Chile) examined. Activation of the RAS/RAF/MAP kinase pathway in biliary tract cancers is likely to be secondary to oncogenic RAS mutations, or due to mutations of the BRAF gene at nucleotide positions not explored in the current study.
Similar content being viewed by others

Main
Biliary tract cancers, which include cancers of the gallbladder, and the intrahepatic and extrahepatic bile ducts, affect over 7500 individuals each year in the United States and many thousands more the world over.1 Biliary tract cancer is a lethal disease, and greater than 50% of affected patients die from their cancer. Complete surgical resection of localized disease is the only avenue for cure—the benefit from postoperative chemotherapy and radiation is minimal in most cases. Therefore, detection of biliary cancer at an early, and hence potentially curable stage, remains crucial for improving survival.2
Currently, the sensitivity and specificity of laboratory tests for preoperative diagnosis of biliary tract cancers is less than optimal, and there are considerable difficulties in distinguishing malignant from benign causes of biliary obstruction. For example, cytologic specimens from brush biopsies have a notorious propensity for yielding false positives and false negatives, with an unacceptable overall sensitivity in the range of only 33–60%.3, 4 Cancer antigen (CA) 19–9 is widely used for serologic detection of biliary cancer, and has a sensitivity of 50–60% and specificity of 80%,5 although others have found it to be a far more unreliable marker of malignancy, especially for the diagnosis of early stage cancers.6 Similarly, detection of p53 and RAS gene mutations in bile has a sensitivity of only 33% and a specificity of 87%.7 There is an ongoing need to identify highly sensitive and specific biomarkers for early detection of biliary tract cancer in biological samples, such as bile or serum.
The v-raf murine sarcoma viral oncogene homolog B1 (BRAF) oncogene encodes a serine/threonine kinase that acts in the MAP kinase pathway, through both receptor tyrosine kinases and G-protein coupled receptors.8 Mutations of BRAF were first reported in melanomas and colorectal cancers, but have since been reported in a variety of solid tumors.9, 10, 11, 12, 13, 14, 15, 16 A 1796 T → A transversion in exon 15 of the BRAF oncogene resulting in a V599E amino-acid missense mutation accounts for at least 80% of the mutations detected in melanomas.8, 12, 14, 17 The BRAFV599E mutant possesses 10-fold greater basal kinase activity than wild-type BRAF, and is the only detected mutation that causes constitutive activation of BRAF independent of RAS activation, by converting BRAF into a dominant transforming protein.8, 18 For cancers harboring BRAF mutations, the exon 15 V599E is by far the most common variant, with a minor proportion of mutations arising in adjacent codons, or in a second cluster on exon 11. In this regard, the BRAF mutational profile bears striking resemblance to the RAS gene, where most cancer-associated mutations are localized to codons 12 or 13. Naturally, this inherent mutation clustering also raises the possibility of developing convenient assays for detecting BRAFV599E as a cancer biomarker in clinical samples. With this motivation, we decided to explore the frequency of the V599E BRAF mutation in a series of archival biliary tract cancers, using three different mutation detection platforms. Our results demonstrate that (a) the V599E mutation is absent in biliary tract cancers arising in the population examined in this study, and therefore unlikely to be a useful cancer detection biomarker, and (b) activation of the RAS/RAF/MAP kinase pathway in biliary tract cancers is likely to be secondary to oncogenic RAS mutations, or due to mutations of the BRAF gene at nucleotide positions not explored in the current study.
Patients and methods
Sample Selection and DNA Isolation
Permission to perform this study was obtained from the Johns Hopkins Joint Committee on Clinical Investigation (JCCI). Archival formalin-fixed paraffin-embedded blocks from 62 surgically excised primary biliary tract cancers were retrieved. The samples included 40 cancers from the Johns Hopkins Hospital surgical pathology archives, including 15 gallbladder cancers, 15 extrahepatic cholangiocarcinomas, and 10 intrahepatic cholangiocarcinomas. The remaining 22 archival cases were gallbladder cancers obtained from Catholic University, Santiago; gallbladder cancers are endemic in the Chilean population, and are the most common cause of cancer-associated deaths in women. The original hematoxylin and eosin (H&E)-stained slides were reviewed by two pathologists (AM, PA) to confirm the diagnosis, and an appropriate block was selected for DNA extraction. Two unstained 10 μm serial sections were obtained, and using the H&E-stained slide as a reference, sections were microdissected using a sterile scalpel under microscopic visualization to obtain greater than 80% neoplastic cells. DNA was extracted using standard protocols as previously published.
PCR Amplification of BRAF Exon 15
PCR primer sequences were designed to amplify a 102 bp fragment of exon 15 (5′-GAA GAC CTC ACA GTA AAA ATA GGT GA-3′, and 5′-CCA CAA AAT GGA TCC AGA CA-3′).13, 19, 20 PCR amplification was performed using 100 ng of tumor sample DNA as template. The PCR reactions were carried out in a 96-well thermocycler. Cycling conditions were as follows: a denaturation step at 95°C for 5 min was followed by two cycles of denaturation at 95°C for 1 min, annealing at 60°C for 1 min, primer extension at 72°C for 1 min, two cycles of denaturation at 95°C for 1 min, annealing at 58°C for 1 min, primer extension at 72°C for 1 min, 35 cycles of denaturation at 95°C for 1 min, annealing at 56°C for 1 min, primer extension at 72°C for 1 min, and one final extension at 72°C for 5 min. Amplified fragments were separated on an agarose gel and visualized by ethidium bromide staining. PCR products were purified using a Qiagen gel extraction kit (Qiagen, Valencia, MA, USA). The PCR products were then divided for further analysis by direct sequencing, Mutector® assay (TrimGen, Sparks, MD, USA), and quantitative Gap Ligase Chain Reaction (GLCR). The technique for direct automated sequencing of BRAF exon 15 has been previously described.13, 19, 20
Mutector® Assay
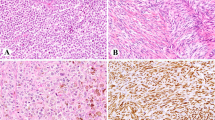
The Mutector® assay is designed for mutation detection for known nucleotide substitutions (http://www.trimgen.com). In brief, a complementary detection primer is designed whose 3′ end terminates at the base immediately preceding the queried nucleotide position (in this case, BRAF T1796). The PCR product from the preceding step is added to wells containing immobilized detection primer and labeled nucleotides. When the target base is wild type, further primer extension does not occur. As a result, the reaction is terminated, labeled nucleotides are not incorporated, and a chromogenic reaction is not observed. When the target base is mutated (eg T → A transversion at BRAF T1796), primer extension continues and a strong chromogenic reaction is observed (Figure 1); notably primer extension will occur with any missense mutation at the queried base (ie, an A, G, or C mutation at T1796 can all elicit a chromogenic reaction). A volume of 10 μl of PCR products of the 102 bp fragment of BRAF exon15 was used as the template for the Mutector® assay. The assay was performed according to the manufacturer's instructions. As a positive control for the BRAF T1796A mutation, we tested the cutaneous melanoma cell line HTB 72. The cervical cancer cell line ME180 served as a negative control.
The Mutector® colorimetric BRAF detection assay is shown in this figure. When the target base is mutated (eg T → A transversion at BRAF T1796), primer extension ensues and a strong color reaction is observed. This is seen in the BRAF-positive control (HTB 72) well (bottom left). The cervical cancer cell line ME180 served as a negative control (top left). Six BRAF negative biliary cancer samples are seen on the right.
Real-Time Quantitative Gap Ligase Chain Reaction (GLCR)
Real-time quantitative GLCR is a sensitive method for detecting point mutations at low level in a background of excess of wild-type DNA. Ligase detection reactions involve the use of two adjacent oligonucleotide primers, which hybridize to a single strand of target DNA, which will be ligated only if there is an exact match to the target sequence.21 In the presence of a mismatch, ligation will not occur. Point mutations are best detected by designing oligonucleotides so that the mutation site is at the 3′ end of the upstream 5′ primer. This linear reaction can increase exponentially by the addition of two further complementary oligonucleotides that hybridize to the perfect-match complementary DNA strand. Using a thermostable ligase in a cycling reaction, ligated products from both reactions are subsequently used as templates, creating exponential amplification, dubbed a Ligase Chain Reaction (LCR). An additional modification to further improve specificity is the use of a single-nucleotide gap at the site of the point mutation between the adjacent oligonucleotide primers. This gap is filled using a thermostable DNA polymerase in the presence of the appropriate mutant nucleotide in the reaction mix, thereby adding substantial additional specificity to the reaction. Based on these principles, we have developed a modified GLCR that can be monitored in real-time using fluorescent-labeled oligonucleotides to allow quantification. In our assay, one oligonucleotide is labeled with a reporter dye at the 5′ end and the adjacent oligonucleotide to which it will ligate, is labeled at the 3′ end with a quencher dye. Excitation of the reporter dye results in fluorescence energy transfer to the quencher dye and subsequent fluorescence at the characteristic spectrum of the quencher dye. The onset of rise in fluorescence by cycle number is proportional to the initial amount of target mutation. DNA from 62 biliary cancers, HTB72 (positive control) and ME180 (negative control) were PCR amplified as stated above. The concentration of all purified products was determined using the TD-360 MiniFluorometer (Turner Designs, Sunnyvale, CA, USA). Stock solutions of each PCR product were diluted in 10 mM Tris pH 8 to yield 1010 amplicon copies/5 μl (calculated from the size of the BRAF 102 bp amplicon assuming 1 bp dsDNA=660 pg/pmol and 1 pmol=6.02 × 1011 copies). For the GLCR assay, 109 copies of PCR product template from each sample were used. In addition, serial 10-fold dilutions (range 1:1–1:10 000) of the positive control (HTB72) mixed into negative control (ME180) were constructed, and 109 copies from each dilution used to generate standard comparison curves against which to compare the unknown biliary cancer samples. Each reaction contained in a 25 μl reaction volume: 2.5 μl Platinum Taq 10 × buffer, 0.625 μl Taq ligase 10 × buffer, 1 mM NADβ, 100 μM mutant-complementary insertion nucleotide, 1 U Platinum Taq polymerase (Invitrogen), 8 U Taq DNA ligase (NEB), 400 nM two nonlabeled reverse-strand mutation-specific oligomers, 400 nM one FAM 5′ labeled forward-strand mutation-specific oligomer and 600 nM one TAMRA 3′ labeled forward-strand mutation-specific oligomer. Sequences for all the oligonucleotides (Operon/Qiagen) are available on request. Reactions were run for 40 cycles (94°C for 2 min initiation, then 50° 30 s, 94°C 30 s) on a 96-well plate using an Applied Biosystems 7700 Sequence detector (Applied Biosystems, CA, USA). All samples were run in duplicate, and were run concurrently on the same plate. Each plate also included multiple water blanks as an additional negative control.
Results
All of the 62 biliary carcinoma samples were negative for the BRAF V599E mutation by all three detection methods. Direct sequencing did not demonstrate any BRAF exon 15 mutations (data not shown). Similarly, using the Mutector® assay, a strong color reaction was observed for the HTB72 positive control; no color reaction was observed for the ME180 negative control or in the biliary cancer samples (Figure 1). Finally, with the GLCR (Figure 2), all 62 biliary cancers demonstrated fluorescence far below the lowest threshold of the standard dilution curve (1:10 000), consistent with a wild-type BRAF gene.
Standard curve for GLCR, generated from dilutions of BRAF-positive DNA into BRAF-negative DNA. A log plot of fluorescence (y-axis) vs PCR cycle (x-axis) is shown. This plot shows T:N serial dilution curves (1:100–1:10 000). All biliary cancer samples in this run are seen far below threshold of the lowest dilution curve.
Discussion
In this study, we failed to find evidence of the common BRAF V599E mutation in 62 archival biliary cancers from all three sites in the biliary tree (gallbladder, intrahepatic and extrahepatic bile ducts). We used three different mutation detection techniques, including quantitative assays like GLCR that would be able to detect the mutation even in the presence of wild-type cells or if present within a subpopulation of tumor cells. While two of the techniques we used (Mutector® and GLCR) only queried BRAF codon 599, the direct sequencing also confirmed absence of mutations in additional codons of exon 15, as has been reported in some cancers. A recent study by Tannapfel et al22 from Germany had reported BRAF mutations in 22% of intrahepatic cholangiocarcinomas; gallbladder or extrahepatic cancers were not examined. The authors performed a more comprehensive direct sequencing analysis of the BRAF gene than in the current study (exons 2–18 were analyzed); nevertheless, of the 15 BRAF mutations identified by the authors, as many as 13 (87%) were localized to exon 15, of which all but two resulted in T1796 alterations. Thus, although we do not detect mutations in the 62 biliary cancers, it is highly unlikely that a substantial proportion is being missed by not examining the entire BRAF gene. We conceded, however, that a small but distinct possibility exists of BRAF mutations occurring in other exons (most importantly, the minor cluster on exon 118) in biliary cancers. Both studies examined primary tumors, and hence an argument for BRAF mutations occurring during disease progression cannot be made. Both studies used microdissected samples; although our tumor enrichment method was not based on the more refined laser capture microdissection, the Mutector® assay can detect an aberrant clonal population up to a 1:150 dilution (see http://www.trimgen.com), while the GLCR technique is even more sensitive, easily detecting a mutation at 1:10 000 dilution (see Figure 2). Thus, it is highly unlikely that our results represent false negatives due to contamination by wild-type cells. A more plausible explanation is that BRAF mutations are rare or absent in biliary tract cancers, at least for those cancers arising on this continent. There may be at least three factors contributing to this discrepancy, the first of which would be geographic factors. Significant differences in molecular abnormalities are common in studies conducted across different continents, even in the same cancer type.23, 24 In this study, we had examined cases from United States and Chile, while Tannapfel's work was based solely on German cases. Secondly, there could be major epidemiological differences between the two sets. For example, it is not stated whether any of the cases examined in the Tannapfel series were related to cholangiocarcinomas arising in the setting of primary sclerosing cholangitis (PSC), while none of the cases in our setting had a known history of PSC. Cholangiocarcinomas arising in the setting of PSC have certain unique genetic abnormalities (eg, p16 promoter mutations, as opposed to the more common p16 CpG island methylation25). Thirdly, attention is drawn to the fact that the Tannapfel report examined only intrahepatic cholangiocarcinomas, while we have examined a mix of cancers from all anatomic portions in the biliary tree (intrahepatic, extrahepatic, and gallbladder carcinomas). If BRAF mutations are critical to the pathogenesis of intrahepatic cholangiocarcinomas but not other biliary tract cancers, we may have missed a positive case based on the relatively small number of intrahepatic tumors (n=15) examined.
Our report is consistent with the emerging evidence that while BRAF mutations are common in certain cancer types, such as colorectal and thyroid cancers and melanomas, they are rare to absent in many other malignancies, such as pancreatic, gastric, renal cell, and head and neck cancers.26, 27, 28, 29, 30 In fact, even within one given cancer type, there exist dramatic differences in frequency of mutations based on tissue origin; thus while 80% of cutaneous melanomas harbor BRAF mutations, these are essentially absent in melanomas of the uveal tract.19, 31 The rarity (or absence) or BRAF mutations is biliary cancers implies that the V599E mutation is unlikely to be a useful biomarker for early detection of these cancers in bile fluids.
References
de Groen PC, Gores GJ, LaRusso NF, et al. Biliary tract cancers. N Engl J Med 1999;341:1368–1378.
Gores GJ . Early detection and treatment of cholangiocarcinoma. Liver Transpl 2000;6:S30–S34.
Mansfield JC, Griffin SM, Wadehra V, et al. A prospective evaluation of cytology from biliary strictures. Gut 1997;40:671–677.
Sturm PD, Rauws EA, Hruban RH, et al. Clinical value of K-ras codon 12 analysis and endobiliary brush cytology for the diagnosis of malignant extrahepatic bile duct stenosis. Clin Cancer Res 1999;5:629–635.
Patel AH, Harnois DM, Klee GG, et al. The utility of CA 19-9 in the diagnoses of cholangiocarcinoma in patients without primary sclerosing cholangitis. Am J Gastroenterol 2000;95:204–207.
Bjornsson E, Kilander A, Olsson R . CA 19-9 and CEA are unreliable markers for cholangiocarcinoma in patients with primary sclerosing cholangitis. Liver 1999;19:501–508.
Muller P, Ostwald C, Puschel K, et al. Low frequency of p53 and ras mutations in bile of patients with hepato-biliary disease: a prospective study in more than 100 patients. Eur J Clin Invest 2001;31:240–247.
Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–954.
Fukushima T, Suzuki S, Mashiko M, et al. BRAF mutations in papillary carcinomas of the thyroid. Oncogene 2003;22:6455–6457.
Xu X, Quiros RM, Gattuso P, et al. High prevalence of BRAF gene mutation in papillary thyroid carcinomas and thyroid tumor cell lines. Cancer Res 2003;63:4561–4567.
Gemignani ML, Schlaerth AC, Bogomolniy F, et al. Role of KRAS and BRAF gene mutations in mucinous ovarian carcinoma. Gynecol Oncol 2003;90:378–381.
Mercer KE, Pritchard CA . Raf proteins and cancer: B-Raf is identified as a mutational target. Biochim Biophys Acta 2003;1653:25–40.
Cohen Y, Xing M, Mambo E, et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst 2003;95:625–627.
Pollock PM, Meltzer PS . A genome-based strategy uncovers frequent BRAF mutations in melanoma. Cancer Cell 2002;2:5–7.
Yuen ST, Davies H, Chan TL, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res 2002;62:6451–6455.
Naoki K, Chen TH, Richards WG, et al. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res 2002;62:7001–7003.
Dong J, Phelps RG, Qiao R, et al. BRAF oncogenic mutations correlate with progression rather than initiation of human melanoma. Cancer Res 2003;63:3883–3885.
Hingorani SR, Jacobetz MA, Robertson GP, et al. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res 2003;63:5198–5202.
Cohen Y, Goldenberg-Cohen N, Parrella P, et al. Lack of BRAF mutation in primary uveal melanoma. Invest Ophthalmol Vis Sci 2003;44:2876–2878.
Singer G, Oldt III R, Cohen Y, et al. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst 2003;95:484–486.
Abravaya K, Carrino JJ, Muldoon S, et al. Detection of point mutations with a modified ligase chain reaction (Gap-LCR). Nucleic Acids Res 1995;23:675–682.
Tannapfel A, Sommerer F, Benicke M, et al. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut 2003;52:706–712.
Biagi JJ, Seymour JF . Insights into the molecular pathogenesis of follicular lymphoma arising from analysis of geographic variation. Blood 2002;99:4265–4275.
Johansson B, Mertens F, Mitelman F . Geographic heterogeneity of neoplasia-associated chromosome aberrations. Genes Chromosomes Cancer 1991;3:1–7.
Taniai M, Higuchi H, Burgart LJ, et al. p16INK4a promoter mutations are frequent in primary sclerosing cholangitis (PSC) and PSC-associated cholangiocarcinoma. Gastroenterology 2002;123:1090–1098.
Ishimura N, Yamasawa K, Karim Rumi MA, et al. BRAF and K-ras gene mutations in human pancreatic cancers. Cancer Lett 2003;199:169–173.
Calhoun ES, Jones JB, Ashfaq R, et al. BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. Am J Pathol 2003;163:1255–1260.
Kim IJ, Park JH, Kang HC, et al. Mutational analysis of BRAF and K-ras in gastric cancers: absence of BRAF mutations in gastric cancers. Hum Genet 2003;114:118–120.
Nagy A, Balint I, Kovacs G . Frequent allelic changes at chromosome 7q34 but lack of mutation of the BRAF in papillary renal cell tumors. Int J Cancer 2003;106:980–981.
Weber A, Langhanki L, Sommerer F, et al. Mutations of the BRAF gene in squamous cell carcinoma of the head and neck. Oncogene 2003;22:4757–4759.
Edmunds SC, Cree IA, Di Nicolantonio F, et al. Absence of BRAF gene mutations in uveal melanomas in contrast to cutaneous melanomas. Br J Cancer 2003;88:1403–1405.
Acknowledgements
This work has been generously supported by the family of Margaret Lee. AM is supported by the Johns Hopkins Clinical Scientist Award and a grant from the Cancer Research and Prevention Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Goldenberg, D., Rosenbaum, E., Argani, P. et al. The V599E BRAF mutation is uncommon in biliary tract cancers. Mod Pathol 17, 1386–1391 (2004). https://doi.org/10.1038/modpathol.3800204
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800204
Keywords
This article is cited by
-
Cholangiocarcinoma with intraductal tubular growth pattern versus intraductal papillary growth pattern
Modern Pathology (2016)
-
BRAF V600E-specific immunohistochemistry reveals low mutation rates in biliary tract cancer and restriction to intrahepatic cholangiocarcinoma
Modern Pathology (2014)
-
Targeted therapy for biliary tract cancers
Journal of Hepato-Biliary-Pancreatic Sciences (2012)
-
Mutational profiling reveals PIK3CA mutations in gallbladder carcinoma
BMC Cancer (2011)