Abstract
Epstein–Barr virus infection has been associated with lymphoepithelioma-like carcinoma of the lung in Asian patients. Recently, Epstein–Barr virus proteins or genomic DNAs were detected in pulmonary squamous-cell carcinoma, adenocarcinoma, and undifferentiated small-cell carcinoma in American patients. We studied 23 cases of small-cell carcinoma of the lung for evidence of Epstein–Barr virus infection by in situ hybridization, immunohistochemistry, and polymerase chain reaction methods. Of the 23 cases, 13 cases were primary small-cell carcinoma of the lung and 10 cases were metastatic small-cell carcinoma of the lung to the brain (one case), liver (two cases), and lymph nodes (seven cases). None of the 23 cases was positive for Epstein–Barr virus-encoded small nonpolyadenylated RNA (EBER)-1 by in situ hybridization. By immunohistochemistry, eight cases showed focal positivity for Epstein–Barr virus nuclear antigen-1. The positive immunostaining was focal and was observed in tumor cells, vascular endothelial cells, and lymphocytes, suggesting nonspecific staining. None of the 23 cases was positive for the transactivating immediate-early BZLF1 (ZEBRA) and latent membrane protein (LMP-1). Only one case was positive for the BamHI W region and LMP-1 gene by polymerase chain reaction assay. Some tumor cells in the BamHI W region positive case were also positive for Epstein–Barr virus nuclear antigen-1. Our study indicates that rare cases of American small-cell carcinoma of the lung may contain Epstein–Barr virus-infected cells, but it is unlikely that Epstein–Barr virus plays a role in the tumorigenesis of small-cell carcinoma of the lung.
Similar content being viewed by others
Main
Epstein–Barr virus has been associated with several human epithelial neoplasms, including nasopharyngeal carcinoma,1 gastric carcinoma, and lymphoepithelioma-like carcinomas from the stomach,2 salivary gland,3 head and neck,4, 5, 6 and gallbladder.7 Pulmonary carcinomas with morphologic features of lymphoepithelioma-like carcinoma have also been reported to harbor Epstein–Barr virus.8, 9, 10, 11 Most of these reports of Epstein–Barr virus-associated epithelial malignancies were in Asian patients. In contrast, there are few reports demonstrating Epstein–Barr virus infection in pulmonary carcinoma in Western populations. Huber et al12 reported a case of pulmonary adenocarcinoma with signet-ring-cell features harboring Epstein–Barr virus by EBER-1 in situ hybridization and BamHI W region polymerase chain reaction methods. Using Epstein–Barr virus nuclear antigen-1 immunohistochemistry, Grinstein et al13 recently found that two of the 22 cases of pulmonary squamous-cell carcinoma and two of the 10 cases of pulmonary neuroendocrine carcinoma were positive for Epstein–Barr virus. Finally, Hartmann et al14 found 11 of the 15 cases of the lung undifferentiated small-cell carcinoma to be positive for Epstein–Barr virus by immunohistochemistry, in situ hybridization and polymerase chain reaction methods.
Small-cell carcinoma of the lung comprises approximately 15% of all pulmonary cancers. At least 85% of patients are smokers. However, only a minority of smokers develop small-cell carcinoma of the lung. Additional factors may play a role in the development of small-cell carcinoma of the lung. As suggested above, Epstein–Barr virus has been proposed to play such a role in the tumorigenesis of small-cell carcinoma of the lung. Therefore, we studied Epstein–Barr virus infection in 23 cases of American small-cell carcinoma of the lung using a large battery of techniques, including in situ hybridization (EBER-1), immunohistochemistry (LAMP-1, ZEBRA, and Epstein–Barr virus nuclear antigen-1) and polymerase chain reaction (LMP-1, Epstein–Barr virus nuclear antigen-4 and BamHI W region) methods.
Materials and methods
Patients and Tissue Samples
In all, 23 cases of small-cell carcinoma of the lung (primary, 13 cases; and metastatic, 10 cases) were retrieved from the surgical pathology files at the Department of Pathology at City of Hope National Medical Center (11 cases) and at the Department of Pathology at the University of Virginia (12 cases) (Table 1). The tissues had been routinely fixed in 10% neutral formalin and embedded in paraffin. One paraffin tissue block with tumor was selected from each case. Several 4 μm sections were cut from each case.
EBER-1 In situ Hybridization
The in situ hybridization methods have been previously described.15 Briefly, we used a probe from a region of the Epstein–Barr virus genome that is actively transcribed in latently infected cells, a 30-base oligonucleotide complementary to a portion (69–98 bp) of the EBER-1 gene. The sequence was 5′-AGA CAC CGT CCT CAC CAC CCG GGA CTT GTA-3′ (Operon Technologies, San Pablo, CA, USA). The probe was labeled with biotin at its 3′ end. Paraffin sections were deparaffinized and digested with pronase (nuclease-free). Sections were incubated with prehybridization solution and then hybridized with sheared salmon sperm and yeast tRNA along with the appropriate amount of probe. The probe was used at a concentration of 0.25 ng/μl with overnight hybridization. Sections were then incubated in a solution of avidin-alkaline phosphatase conjugate, washed for 3 min, incubated in McGadey's substrate, briefly washed in distilled water, air dried, and coverslipped. No counterstain was used. A poly-d(T) was used as a control for total RNA preservation, while a known Epstein–Barr virus-positive case of nasopharyngeal carcinoma was used as a positive control. A case was considered positive if the nucleus, or the nucleus and cytoplasm, of a tumor cell stained dark blue or black.
Epstein–Barr Virus Nuclear Antigen-1, LMP-1, and ZEBRA Immunohistochemistry
Dr Grasser16 (Abteilung Virologie, Institut fur Medizinische Mikrobiologie und Hygiene and Institut fur Pathologie, Universitaatakliniken des Saarlandes, Homburg, Germany) kindly provided the rat monoclonal antibody clone 2B4 to Epstein–Barr virus nuclear antigen-1. We also used two other antibodies, mouse monoclonal antibody clones CS1-4 to LMP-1 protein (Dako Corporation, Carpinteria, CA, USA) and clone BZ.1 to ZEBRA (BamHI Z fragment, Epstein–Barr-replication activator) protein (Dako). Epstein–Barr virus nuclear antigen-1, LMP-1, and ZEBRA immunohistochemistry were performed in all 23 cases of small-cell carcinoma of the lung. Paraffin sections were deparaffinized and rehydrated in a graded alcohol series. Two of the antibodies (LMP-1 and ZEBRA) required heat-induced epitope retrieval using 100 mM EDTA buffer (pH 8.0) or 10 mM citrate buffer (pH 6.0), for Epstein–Barr virus nuclear antigen-1 and ZEBRA, respectively, in a steamer (Black and Decker, Shelton, CT, USA) at 100°C for 20 min. The sections were then incubated with 2B4 at 1:500 dilution at room temperature overnight, with CS1-4 at 1:320, or with BZ.1 at 1:20 dilution at room temperature for 40 min and washed three times (5 min each) with PBS buffer. The sections were then incubated with a biotinylated goat, anti-rat antibody (Vector Laboratories, Burlingame, CA, USA) (for Epstein–Barr virus nuclear antigen-1) at a dilution of 1:150, or biotinylated goat, anti-mouse/anti-rabbit antibody (Ventana Medical Systems, Tucson, AZ, USA) (for LMP-1 and ZEBRA) at a dilution of 1:8, followed by the application of two washes (5 min each) of PBS buffer. EnVision Plus (Dako) and peroxidase detection methods were used.
Polymerase Chain Reaction Studies for LMP-1, Epstein–Barr Virus Nuclear Antigen-4 and BamHI W Region
Viral genomic DNA was extracted from formalin-fixed, paraffin-embedded tumor tissues, using 0.2 mg/ml. proteinase K digestion buffer overnight, followed by denaturation by boiling. The polymerase chain reaction studies were performed with 2 μl of extracted DNA in a 30 μl mixture containing 50 mmol/l KCl, 10 mmol/l Tris buffer (pH 8.3), 50 μm of each deoxynucleotide triphosphate, 2.5 mmol/l MgCl2, 1 U of Taq polymerase (Perkin Elmer, Foster City, CA, USA) and 20 pmol of each primer. The primers for BamHI W region were: 3′-CGGTCGCCCAGTCCTACCAG-5′ and 5′-CCTGGAGAGGTCAGGTTACT-3′.17 The expected BamHI W region PCR product size was 125 bp. We used primers for Epstein–Barr virus nuclear antigen-4 that flank the DNA region coding for epitopes of 399–408 and 416–424 of the prototype B95.8 Epstein–Barr virus: Epstein–Barr virus nuclear antigen-4+ 5′-GAG GAG GAA GAC AAG AGT GG-3′ and Epstein–Barr virus nuclear antigen-4− 5′-GAT TCA GGC GTG GCT CTT GG-3′. We also used primers for the Epstein–Barr virus LMP-1 gene that flank the site of the characteristic 30 bp deletion of LMP-1 gene: LMP-1+ 5′-CGG AAG AGG TGG AAA ACA AA-3′ and LMP-1− 5′-GTG GGG GTC GTC ATC ATC TC-3′. After initial denaturation for 5 min at 95°C, 45 amplification cycles were performed as follows: denaturing at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 40 s. A final extension at 72°C for 7 min completed the polymerase chain reaction amplification. The polymerase chain reaction setup and the post-polymerase chain reaction work were performed in separate laboratories to minimize the possibility of contamination. The BamHI W, Epstein–Barr virus LMP-1, and Epstein–Barr virus nuclear antigen-4 polymerase chain reaction products were separated by agarose gel electrophoresis and transferred on to a nitrocellulose membrane. The BamHI W region polymerase chain reaction product was then hybridized with BamHI W region-specific probe 5′-GGGGACTTCCACTTGGCGAA-3′. The LMP-1 polymerase chain reaction product was hybridized with LMP-1 probe 5′- CCGGAACCAGAAGTACCCAA-3′. The Epstein–Barr virus nuclear antigen-4 polymerase chain reaction product was hybridized with Epstein–Barr virus nuclear antigen-4 probe 5′- ACTGCCGTACAATCCAACAG-3′.
Results
Clinical Features
All 23 patients were Caucasians. Of the 23 cases, 18 were resection specimens, and five were needle-core biopsy specimens.
In situ Hybridization for EBER-1
Sections of all 23 cases of small-cell carcinoma of the lung were hybridized with poly-d(T) and EBER-1. All the cases showed strong nuclear positivity for poly-d(T) (control for RNA preservation). None of the 23 cases was positive for EBER-1 (Figure 1, Table 1).
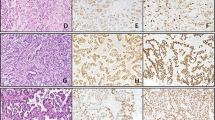
Liver metastatic small-cell carcinoma of the lung (case 8) had amplified BamHI W region product by polymerase chain reaction (top). The amplified 125 bp product in lane 1 corresponds the size of control Raji cells (lane 8). All other six cases (lanes 2–7) are negative. By immunohistochemistry, many tumor cells were positive for Epstein–Barr virus nuclear antigen-1 by 2B4 immunohistochemistry (lower left). However, none of tumor cells was positive for EBER-1 by in situ hybridization (lower right). In addition, polymerase chain reaction amplified LMP-1 and Epstein–Barr virus nuclear antigen-4 products were not identified in this case. Immunostains for LMP-1 and ZEBRA were also negative.
Immunohistochemical Studies for Epstein–Barr Virus Nuclear Aantigen-1, ZEBRA and LMP-1
Of the 23 cases of small-cell carcinoma of the lung eight were positive for Epstein–Barr virus nuclear antigen-1, showing a granular nuclear staining in tumor cells (Figure 2a), lymphocytes (Figure 2b), or vascular endothelial cells (Figure 2c). Some tumor cells also showed cytoplasmic staining (Figure 2d). The percentage of Epstein–Barr virus nuclear antigen-1 positive tumor cells varied from less than 1% to more than 10%. None of the 23 cases was positive for ZEBRA or LMP-1 protein.
Nonspecific 2B4 immunostains seen in small-cell carcinoma of the lung. (a) Shows nuclear stain in tumor cells; (b) shows positive nuclear stain in infiltrating lymphoid cells; (c) shows rare vascular endothelial cells being nuclear positive for 2B4; (d) shows clusters of tumor cell with cytoplasmic 2B4 stain.
LMP-1, Epstein–Barr Virus Nuclear Antigen-4 and BamHI W Internal Repeat Region Polymerase Chain Reaction Studies
DNA from formalin-fixed and paraffin-embedded tissues of all the 23 cases were successfully purified and amplified with primers flanking BamHI W region, LMP-1, Epstein–Barr virus nuclear antigen-4 and β-globulin (the latter, a control for DNA preservation). Strong β-globin amplified bands were identified from all the 23 cases, indicating that the quality and quantity of the purified DNAs were good. One of the 23 cases showed an amplified BamHI W region (Figure 1). Some tumor cells in this case had shown nuclear positivity for 2B4 by immunohistochemistry, but was negative by other assays (Figure 1). One other case was polymerase chain reaction positive for the LMP-1 gene region. However, this case was negative by all other assays. None of the 23 cases was positive for the Epstein–Barr virus nuclear antigen-4 gene by polymerase chain reaction.
Discussion
Several studies have demonstrated Epstein–Barr virus genome in pulmonary carcinomas with morphologic features of lymphoepithelioma-like carcinoma in Asian pattents8, 9, 10, 11 and Western populations. 12 We studied Epstein–Barr virus RNA in 78 cases of pulmonary nonlymphoepithelioma-like carcinoma using in situ hybridization methodology in 1992, and found none of them to be positive for Epstein–Barr virus.15 In contrast, several recent studies reported that Epstein–Barr virus gene products were detected in pulmonary nonlymphoepithelioma-like carcinoma, such as pulmonary squamous cell carcinoma and neuroendocrine carcinoma13 and small-cell carcinoma of the lung.14 It is necessary to use a large battery of molecular and immunohistochemical methods to investigate whether Epstein–Barr virus plays any role in pulmonary nonlymphoepithelioma-like carcinoma.
EBER in situ hybridization has been regarded as ‘gold standard’ for the identification of a significant association of Epstein–Barr virus with a given neoplasm.18 Unlike Epstein–Barr virus nuclear antigens and LMPs, EBERs are theoretically expressed in all three types of Epstein–Barr virus latent infection. Epstein–Barr virus-infected cells usually contain a large number of EBER copies, on the order of 105–106, therefore EBER in situ hybridization should be the most sensitive method to detect any type of Epstein–Barr virus infection in tumor cells. Most importantly, by direct visualization of positive cells, one can determine whether tumor cells or normal lymphocytes are positive for Epstein–Barr virus. However, several recent studies have cast doubt on the ability of EBER in situ hybridization studies,19, 20, 21 although other subsequent studies have not confirmed this finding.22, 23, 24
In the current study, we also performed polymerase chain reaction analysis using two separate target regions. The BamHI W region is iterated 7–12 times in the Epstein–Barr virus genome, thus providing a particularly good target for the detection of Epstein–Barr virus in a sample in which a small viral copy number might be expected.25 However, visualization of the positive cells is not possible using polymerase chain reaction analysis. Approximately one in 105–106 of the B lymphocytes purified from the peripheral blood of previously infected people are latently infected with Epstein–Barr virus.26, 27 Other studies of normal lymphoid tissues have shown between one in 1000 to one in 10000 infected cells.28 When DNA is extracted from tissue for polymerase chain reaction studies, Epstein–Barr virus DNA from Epstein–Barr virus infected B-cells may be potentially amplified, leading to a ‘false’-positive polymerase chain reaction result. Therefore, polymerase chain reaction studies alone do not provide definitive evidence of Epstein–Barr virus within neoplastic cells, as one cannot exclude the possibility that Epstein–Barr virus within ‘bystander’ lymphocytes contributes the positive signal.
Immunohistochemical demonstration of Epstein–Barr virus nuclear antigen-1 expression may provide additional evidence that a given tumor is Epstein–Barr virus positive. Similar to other studies in the literature,13, 19 we also used Epstein–Barr virus nuclear antigen-1 antibody 2B4 clone in our study, which was produced against a lambda-cro/lacZ- Epstein–Barr virus nuclear antigen-1 fusion protein that had been shown to react in all forms of Epstein–Barr virus latency.16 This antibody had been previously demonstrated to have nearly equal sensitivity in paraffin or frozen section immunohistochemistry. However, the current study as well as some of the previous studies have suggested that the specificity of the 2B4 clone may not be absolute.23, 29 The irregular GGGGCAGGA repeat motifs of the latent infection cycle Epstein–Barr virus nuclear antigen-1 gene are interspersed in cellular DNA. Monoclonal antibodies to the irregular glu and ala repeating peptide motifs of Epstein–Barr virus nuclear antigen-1 may theoretically cross-react with cell protein.30 Moreover, two cell proteins can specifically bind to the Epstein–Barr virus nuclear antigen-1 cognate DNA sequence.31 Thus, at least part of the Epstein–Barr virus nuclear antigen-1 reactivity with antibodies may have arisen from this cellular DNA interaction. Therefore, at present, we do not consider Epstein–Barr virus nuclear antigen-1 immunohistochemical positivity as a sole finding as evidence of Epstein–Barr virus infection, in the absence of other confirmatory studies, in particular EBER in situ hybridization. We think that at this time, at least two different techniques, including at least one providing cellular localization to the tumor cells should be found to be positive, before one can consider that a particular neoplasm may be truly Epstein–Barr virus associated.
Of the 23 cases of American small-cell carcinoma of the lung studied by us, we found nine cases with Epstein–Barr virus positivity by at least one technique. Seven of these cases were positive by Epstein–Barr virus nuclear antigen-1 staining alone, without confirmation using a second methodology. In these cases, the percentage of positive tumor cells was low and there was an additional staining of endothelial cells and lymphocytes. We consider this to be false-positive staining since polymerase chain reaction analysis was completely negative in seven of the eight cases, and in the eighth case, only the highly sensitive BamHI W PCR was positive, possibly reflecting ‘contamination’ by Epstein–Barr virus infected B-lymphocytes. We did not find any case to be positive by EBER-1 in situ hybridization, including either the BamHI W region positive case (case 8) or the LMP-1-positive case (case 9). Eight cases with a variable number of tumor cells being positive for Epstein–Barr virus nuclear antigen-1 stain were interpreted as false-positive (nonspecific staining) because a similar Epstein–Barr virus nuclear antigen-1 nuclear staining was also found in vascular endothelial cells and lymphocytes in same cases and the positive Epstein–Barr virus nuclear antigen-1 staining did not show any correlation with the results of EBER-1 in situ hybridization. There was only one case (case 8) that had Epstein–Barr virus nuclear antigen-1 positive cells and was positive with another technique (polymerase chain reaction for the BamHI W region), and this case was negative by all other analyses, including two other polymerase chain reaction assays. We believe that the BamHI W region positivity by polymerase chain reaction in this case most likely represents contamination by host lymphocytes that was detected due to the exquisite sensitivity of the assay.
In summary, after studying 23 cases of small-cell carcinoma of the lung using most sensitive and specific methods, we did not find convincing evidence of Epstein–Barr virus infection. Although some cases of small-cell carcinoma of the lung may be positive, Epstein–Barr virus by polymerase chain reaction and some cells of small-cell carcinoma of the lung may be positive for Epstein–Barr virus nuclear antigen-1 by immunohistochemical assays, these results in isolation are not sufficient to conclude pathogenetic significance. Epstein–Barr virus nuclear antigen-1 immunohistochemistry or polymerase chain reaction assay alone cannot be used as the only method to determine Epstein–Barr virus status in a given tumor.
References
Iezzoni JC, Gaffey MJ, Weiss LM . The role of Epstein–Barr virus in lymphoepithelioma-like carcinoma. Am J Clin Pathol 1995;103:308–315.
Shibata D, Tokunaga M, Uemura Y, et al. Association of Epstein–Barr virus with undifferentiated gastric carcinoma with intense lymphoid infiltration. Am J Pathol 1991;139:469–474.
Huo T, Hsueh C . Lymphoepithelioma-like salivary gland carcinoma in Taiwan: a clinicopathological study of nine cases demonstrating a strong association with Epstein–Barr virus. Histopathology 1997;31:75–82.
Leung SY, Yuen ST, Ho CM, et al. Presence of Epstein–Barr virus in lymphoepithelioma-like carcinoma of the middle ear. J Clin Pathol 1998;51:602–605.
Shimakage M, Horii K, Tempaku A, et al. Association of Epstein–Barr virus with oral cancers. Hum Pathol 2002;33:608–614.
Jeng YM, Sung MT, Fang CL, et al. Sinonasal undifferentiated carcinoma and nasopharyngeal-type undifferentiated carcinoma: two clinically, biologically, and histopathologically distinct entities. Am J Surg Pathol 2002;26:371–376.
Jeng YM, Chen CL, Hsu HC . Lymphoepithelioma-like cholangiocarcinoma: an Epstein–Barr virus-associated tumor. Am J Surg Pathol 2001;25:516–520.
Wong MP, Chung LP, Yuen ST, et al. In situ detection of Epstein–Barr virus in non-small-cell lung carcinomas. J Pathol 1995;177:233–240.
Han AJ, Xiong M, Zong YS . Association of Epstein–Barr virus with lymphoepithelioma-like carcinoma of the lung in southern China. Am J Clin Pathol 2000;114:220–226.
Chen FF, Yan JJ, Lai WW, et al. Epstein–Barr virus-associated nonsmall-cell lung carcinoma: undifferentiated ‘lymphoepithelioma-like’ carcinoma as a distinct entity with better prognosis. Cancer 1998;82:2334–2342.
Curcio LD, Cohen JS, Grannis. Jr FW, et al. Primary lymphoepithelioma-like carcinoma of the lung in a child. Report of an Epstein–Barr virus-related neoplasm. Chest 1997;111:250–251.
Huber M, Pavlova B, Muhlberger H, et al. Detection of the Epstein–Barr virus in primary adenocarcinoma of the lung with signet-ring cells. Virchows Arch 2002;441:25–30.
Grinstein S, Preciado MV, Gattuso P, et al. Demonstration of Epstein–Barr virus in carcinomas of various sites. Cancer Res 2002;62:4876–4878.
Hartmann DP, Luka J, Azumi N . Detection of Epstein–Barr virus (EBV) in small-cell undifferentiated carcinoma of the lung (abstract). Mod Pathol 2001;14:220A.
Chang KL, Chen YY, Shibata D, et al. Description of an in situ hybridization methodology for detection of Epstein–Barr virus RNA in paraffin-embedded tissue, with a survey of normal and neoplastic tissues. Diagn Mol Pathol 1992;1:246–255.
Grasser FA, Murray PG, Kremmer E, et al. Monocolonal antibodies directed against the Epstein–Barr virus-encoded nuclear antigen 1 (EBNA1): immunohistologic detection of EBNA1 in the malignant cells of Hodgkin's disease. Blood 1994;84:3792–3798.
Imai S, Usui N, Sugiura M, et al. Epstein–Barr virus genomic sequences and specific antibodies in cerebrospinal fluid in children with neurologic complications of acute and reactivated EBV infections. J Med Virol 1993;40:278–284.
Ambinder R, Mann R . Epstein–Barr-encoded RNA in situ hybridization. Hum Pathol 1994;25:602–605.
Bonnet M, Guinebretiere JM, Kremmer E, et al. Detection of Epstein–Barr virus in invasive breast cancers. J Natl Cancer Inst 1999;91:1376–1381.
Sugawara Y, Mizugaki Y, Uchida T, et al. Detection of Epstein–Barr virus (EBV) in hepatocellular carcinoma tissue: a novel EBV latency characterized by the absence of EBV-encoded small RNA expression. Virology 1999;256:196–202.
Gan YJ, Razzouk BI, Su T, et al. A defective, rearranged Epstein–Barr virus genome in EBER-negative and EBER-positive Hodgkin's disease. Am J Pathol 2002;160:781–786.
Chu PG, Chen YY, Chen W, et al. No direct role for Epstein–Barr virus in American hepatocellular carcinoma. Am J Pathol 2001;159:1287–1292.
Chu PG, Chang KL, Chen YY, et al. No significant association of Epstein–Barr virus infection with invasive breast carcinoma. Am J Pathol 2001;159:571–578.
Herrmann K, Niedobitek G . Epstein–Barr virus-associated carcinomas: facts and fiction. J Pathol 2003;199:140–145.
Labrecque LG, Barnes DM, Fentiman IS, et al. Epstein–Barr virus in epithelial cell tumors: a breast cancer study. Cancer Res 1995;55:39–45.
Miyashita EM, Yang B, Lam KM, et al. A novel form of Epstein–Barr virus latency in normal b-cells in vivo. Cell 1995;80:593–601.
Yao QY, Rickinson AB, Epstein MA . A re-examination of the Epstein–Barr virus carrier state in healthy seropositive individuals. Int J Cancer 1985;35:35–43.
Deamant FD, Albujar PF, Chen YY, et al. Epstein–Barr virus distribution in nonneoplastic lymph nodes. Mod Pathol 1993;6:729–732.
Brink AATP, van den Brule AJC, van Diest P, Meijer CJLM . Re: Detection of Epstein–Barr virus in invasive breast cancers. J Natl Cancer Inst 2000;92:655–656.
Luka J, Kreofsky T, Pearson GR, et al. Identification and characterization of a cellular protein that cross-reacts with the Epstein–Barr virus nuclear antigen. J Virol 1984;52:833–838.
Wen LT, Lai PK, Bradley G, et al. Interaction of Epstein–Barr viral (EBV) origin of replication (oriP) with EBNA-1 and cellular anti-EBNA-1 proteins. Virology 1990;178:293–296.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chu, P., Cerilli, L., Chen, YY. et al. Epstein–Barr virus plays no role in the tumorigenesis of small-cell carcinoma of the lung. Mod Pathol 17, 158–164 (2004). https://doi.org/10.1038/modpathol.3800024
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800024
Keywords
This article is cited by
-
Sudden onset of paraplegia secondary to an unusual presentation of pediatric synovial sarcoma
Child's Nervous System (2016)
-
Epstein–Barr virus microRNAs and lung cancer
British Journal of Cancer (2011)