Abstract
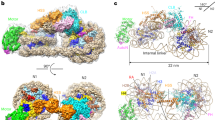
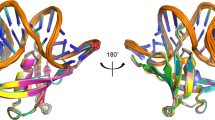
The structure of a complex containing the homeodomain repressor protein MATα2 and the MADS-box transcription factor MCM1 bound to DNA has been determined by X-ray crystallography at 2.25 Å resolution. It reveals the protein–protein interactions responsible for cooperative binding of MATα2 and MCM1 to DNA. The otherwise flexible amino-terminal extension of the MATα2 homeodomain forms a β-hairpin that grips the MCM1 surface through parallel β-strand hydrogen bonds and close-packed, predominantly hydrophobic, side chains. DNA bending induced by MCM1 brings the two proteins closer together, facilitating their interaction. An unusual feature of the complex is that an eight-amino-acid sequence adopts an α-helical conformation in one of two copies of the MATα2 monomer and a β-strand conformation in the other. This ‘chameleon’ sequence of MATα2 may be important for recognizing natural operator sites.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gehring, W. J. et al. Homeodomain-DNA recognition. Cell 78, 211–223 (1994).
Wilson, D. S. & Desplan, C. Homeodomain proteins. Cooperating to be different. Curr. Biol. 5, 32–34 (1995).
Wolberger, C. Homeodomain interactions. Curr. Opin. Struct. Biol. 6, 62–68 (1996).
Vershon, A. K. Protein interactions of homeodomain proteins. Curr. Opin. Biotechnol. 7, 392–396 (1996).
Johnson, A. D. Molecular mechanisms of cell-type determination in budding yeast. Curr. Opin. Genet. Dev. 5, 552–558 (1995).
Sauer, R. T., Smith, D. L. & Johnson, A. D. Flexibility of the yeast alpha 2 repressor enables it to occupy the ends of its operator, leaving the center free. Genes Dev. 2, 807–816 (1988).
Hall, M. N. & Johnson, A. D. Homeodomain of the yeast repressor alpha 2 is a sequence-specific DNA-binding domain but is not sufficient for repression. Science 237, 1007–1012 (1987).
Komachi, K., Redd, M. J. & Johnson, A. D. The WD repeats of Tup1 interact with the homeodomain protein alpha 2. Genes Dev. 8, 2857–2867 (1994).
Wolberger, C., Vershon, A. K., Liu, B., Johnson, A. D. & Pabo, C. O. Crystal structure of a MAT alpha 2 homeodomain–operator complex suggests a general model for homeodomain–DNA interactions. Cell 67, 517–528 (1991).
Phillips, C. L., Vershon, A. K., Johnson, A. D. & Dahlquist, F. W. Secondary structure of the homeodomain of yeast alpha 2 repressor determined by NMR spectroscopy. Genes. Dev. 5, 764–772 (1991).
Mak, A. & Johnson, A. D. The carboxy-terminal tail of the homeodomain protein alpha 2 is required for function with a second homeodomain protein. Genes Dev. 7, 1862–1870 (1993).
Li, T., Stark, M. R., Johnson, A. D. & Wolberger, C. Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA. Science 270, 262–269 (1995).
Lydall, D., Ammerer, G. & Nasmyth, K. Anew role for MCM1 in yeast: cell cycle regulation of SW15 transcription. Genes Dev. 5, 2405–2519 (1991).
Passmore, S., Maine, G. T., Elble, R., Christ, C. & Tye, B. K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MAT alpha cells. J. Mol. Biol. 204, 593–606 (1988).
Kuo, M. H., Nadeau, E. T. & Grayhack, E. J. Multiple phosphorylated forms of the Saccharomyces cerevisiae Mcm1 protein include an isoform induced in response to high salt concentrations. Mol. Cell. Biol. 17, 819–832 (1997).
Althoefer, H., Schleiffer, A., Wassmann, K., Nordheim, A. & Ammerer, G. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 5917–5928 (1995).
Maher, M., Cong, F., Kindelberger, D., Nasmyth, K. & Dalton, S. Cell cycle-regulated transcription of the CLB2 gene is dependent on Mcm1 and a ternary complex factor. Mol. Cell. Biol. 15, 3129–3137 (1995).
Christ, C. & Tye, B. K. Functional domains of the yeast transcription/replication factor MCM1. Genes Dev. 5, 751–763 (1991).
Primig, M., Winkler, H. & Ammerer, G. The DNA binding and oligomerization domain of MCM1 is sufficient for its interaction with other regulatory proteins. EMBO J. 10, 4209–4218 (1991).
Shore, P. & Sharrocks, A. D. The MADS-box family of transcription factors. Eur. J. Biochem. 229, 1–13 (1995).
Pellegrini, L., Tan, S. & Richmond, T. J. Structure of serum response factor core bound to DNA. Nature 376, 490–498 (1995).
Smith, D. L. & Johnson, A. D. Amolecular mechanism for combinatorial control in yeast: MCM1 protein sets the spacing and orientation of the homeodomains of an alpha 2 dimer. Cell 68, 133–142 (1992).
Vershon, A. K. & Johnson, A. D. Ashort, disordered protein region mediates interactions between the homeodomain of the yeast alpha 2 protein and the MCM1 protein. Cell 72, 105–112 (1993).
Keleher, C. A., Goutte, C. & Johnson, A. D. The yeast cell-type specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell 53, 927–936 (1988).
Keleher, C. A., Passmore, S. & Johnson, A. D. Yeast repressor alpha 2 binds to its operator cooperatively with yeast protein Mcm1. Mol. Cell. Biol. 9, 5228–5230 (1989).
Mead, J., Zhong, H., Acton, T. B. & Vershon, A. K. The yeast alpha2 and Mcm1 proteins interact through a region similar to a motif found in homeodomain proteins of higher eukaryotes. Mol. Cell. Biol. 16, 2135–2143 (1996).
Minor, D. L. J & Kim, P. S. Context-dependent secondary structure formation of a designed protein sequence. Nature 380, 730–734 (1996).
Wynne, J. & Treisman, R. SRF and MCM1 have related but distinct DNA binding specificities. Nucleic Acids Res. 20, 3297–3303 (1992).
Acton, T. B., Zhong, H. & Vershon, A. K. DNA-binding specificity of Mcm1: operator mutations that alter DNA-bending and transcriptional activities by a MADS box protein. Mol. Cell. Biol. 17, 1881–1889 (1997).
Zhong, H. L. & Vershon, A. K. The yeast homeodomain protein Mat alpha 2 shows extended DNA binding specificity in complex with Mcm1. J. Biol. Chem. 272, 8402–8409 (1997).
Labeots, L. A. & Weiss, M. A. Electrostatics and hydration at the homeodomain DNA interface: chemical probes of an interfacial water cavity. J. Mol. Biol. 269, 113–128 (1997).
Bruhn, L. & Sprague, G. F. J MCM1 point mutants deficient in expression of alpha-specific genes: residues important for interaction with alpha 1. Mol. Cell. Biol. 14, 2534–2544 (1994).
Abel, K., Yoder, M. D., Hilgenfeld, R. & Jurnak, F. An alpha to beta conformational switch in EF-Tu. Structure 4, 1153–1159 (1996).
Polekhina, G. et al. Helix unwinding in the effector region of elongation factor EF-Tu–GDP. Structure 4, 1141–1151 (1996).
Wright, H. T. The structural puzzle of how serpin serine proteinase inhibitors work. BioEssays 18, 453–464 (1996).
Perutz, M. F. Amyloid fibrils. Mutations make enzyme polymerize. Nature 385, 773–775 (1997).
Navaza, J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994).
Brünger, A. X-PLOR v3.1 Manual(Yale Univ. Press, New Haven, CT, (1992)).
Tronrud, D. E. Conjugate-direction minimization: an improved method for the refinement of macromolecules. Acta Crystallogr. A 48, 912–916 (1992).
Ferrin, T. E., Huang, C. C., Jarvis, L. E. & Langridge, R. The MIDAS display system. J. Mol. Graph. 6, 13–27 (1988).
Lavery, R. & Sklenar, H. The definition of generalised helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dynam. 6, 63–91 (1988).
Nicholls, A., Sharp, K. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Acknowledgements
We thank Y. Hunziker, L. Pellegrini and D. Sargent for their support and assistance, and A. Thompson at BM14 of the ESRF Grenoble for help during data collection. This research was supported in part by the Swiss National Science Foundation.
Author information
Authors and Affiliations
Supplementary Information
Rights and permissions
About this article
Cite this article
Tan, S., Richmond, T. Crystal structure of the yeast MATα2/MCM1/DNA ternary complex. Nature 391, 660–666 (1998). https://doi.org/10.1038/35563
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35563
This article is cited by
-
Design of orthogonal regulatory systems for modulating gene expression in plants
Nature Chemical Biology (2020)
-
Cloning and functional analysis of the flowering gene GmSOC1-like, a putative SUPPRESSOR OF OVEREXPRESSION CO1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in soybean
Plant Cell Reports (2013)
-
Mcm1p binding sites in the ARG1 promoter positively regulate ARG1 transcription and S. cerevisiae growth in the absence of arginine and Gcn4p
Amino Acids (2011)
-
Global mapping of protein-DNA interactions in vivo by digital genomic footprinting
Nature Methods (2009)
-
The origin of protein interactions and allostery in colocalization
Nature (2007)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.