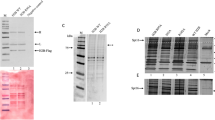
Abstract
The Rho-family of GTPases and their regulators are essential for cytoskeletal reorganization and transcriptional activation in response to extracellular signals1,2. Little is known about what links these molecules to membrane receptors. In the budding yeast Saccharomyces cerevisiae, haploid cells respond to mating pheromone through a G-protein-coupled receptor and the βγ subunit of the G protein3, resulting in arrest of the cell cycle, transcriptional activation, and polarized growth towards a mating partner4,5. The Rho-family GTPase Cdc42 and its exchange factor Cdc24 have been implicated in the mating process6,7, but their specific role is unknown. Here we report the identification of cdc24 alleles that do not affect vegetative growth but drastically reduce the ability of yeast cells to mate. When exposed to mating pheromone, these mutants arrest growth, activate transcription, and undergo characteristic morphological and actin-cytoskeleton polarization. However, the mutants are unable to orient towards a pheromone gradient, and instead position their mating projection adjacent to their previous bud site. The mutants are specifically defective in the binding of Cdc24 to the G-protein βγ subunit. Our results demonstrate that the association of an exchange factor and the βγ subunit of a hetero-trimeric G protein links receptor-mediated activation to oriented cell growth.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Machesky, L. M. & Hall, A. Rho: a connection between membrane receptor signaling and the cytoskeleton. Trends Cell Biol. 6, 304–310 (1996).
Cerione, R. A. & Zheng, Y. The Dbl family of oncogenes. Curr. Opin. Cell Biol. 8, 216–222 (1996).
Whiteway, M. et al. the STE4 and STE18 genes of yeast encode potential β-subunits and γ-subunits of the mating factor receptor-coupled G protein. Cell 56, 467–477 (1989).
Sprague, G. F. J & Thorner, J. W. in The Molecular and Cellular Biology of the Yeast Saccharomyces (eds Jones, E. W., Pringle, J. R. & Broach, J. R.) 657–744 (Cold Spring Harbor Laboratory Press, NY, (1992)).
Leberer, E., Thomas, D. Y. & Whiteway, M. Pheromone signalling and polarized morphogenesis in yeast. Curr. Opin. Genet. Dev. 7, 59–66 (1997).
Simon, M.-N. et al. Role for the Rho-family GTPase Cdc42 in yeast mating pheromone signal pathway. Nature 376, 702–705 (1995).
Zhao, Z. S., Leung, T., Manser, E. & Lim, L. Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol. Cell. Biol. 15, 5246–5257 (1995).
Sloat, B. F., Adams, A. & Pringle, J. R. Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J. Cell. Biol. 89, 395–405 (1981).
Adams, A. E., Johnson, D. I., Longnecker, R. M., Sloat, B. F. & Pringle, J. R. CDC42 and CDC43, two additional genes invovled in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell. Biol. 111, 131–142 (1990).
Chenevert, J., Valtz, N. & Herskowitz, I. Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics 136, 1287–1296 (1994).
Trueheart, J., Boeke, J. D. & Fink, G. R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol. Cell. Biol. 7, 2316–2328 (1987).
Segall, J. E. Polarization of yeast cells in spatial graidents of α mating factor. Proc. Natl Acad. Sci. USA 90, 8332–8336 (1993).
Dorer, R., Pryciak, P. M. & Hartwell, L. H. Saccharomyces cerevisiae cells execute a default pathway to select a mate in the absence of pheromone gradients. J. Cell Biol. 131, 845–861 (1995).
Valtz, N., Peter, M. & Herskowitz, I. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J. Cell Biol. 131, 863–873 (1995).
Zheng, Y., Cerione, R. & Bender, A. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J. Biol. Chem. 269, 2369–2372 (1994).
Zheng, Y., Bender, A. & Cerione, R. A. Interactions among proteins involved in bud-site selection and bud-site assembly in Saccharomyces cerevisiae. J. Biol. Chem. 270, 626–630 (1995).
Chenevert, J., Corrado, K., Bender, A., Pringle, J. & Herskowitz, I. Ayeast gene (BEM1) necessary for cell polarization whose product contains two SH3 domains. Nature 356, 77–79 (1992).
Hirschman, J. E., DeZutter, G. S., Simonds, W. F. & Jenness, D. D. The Gβγ complex of the yeast pheromone response pathway–subcellular fractionation and protein–protein interactions. J. Biol. Chem. 272, 240–248 (1997).
Peter, M., Neiman, A. M., Park, H. O., Van Lohuizen, M. & Herskowitz, I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 15, 7046–7059 (1996).
Leberer, E. et al. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 16, 83–97 (1997).
Rose, M. D., Winston, F. & Hieter, P. Methods in Yeast Genetics: A Laboratory Course Manual (Cold Spring Harbor Laboratory Press, NY, (1991)).
Miyamoto, S. et al. ADBL-homologous region of the yeast CLS4/CDC24 gene product is important for Ca(2+)-modulated bud assembly. Biochem. Biophys. Res. Commun. 181, 604–610 (1991).
Mitchell, D. A., Marshall, T. K. & Deschenes, R. J. Vectors for the inducible overexpression of glutathione-S-transferase fusion proteins in yeast. Yeast 9, 715–722 (1993).
Wach, A., Brachat, A., Alberti-Segui, C., Rebischung, C. & Philippsen, P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13, 1065–1075 (1997).
Fromant, M., Blanquet, S. & Plateau, P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain-reaction. Anal. Biochem. 224, 347–353 (1995).
Sprague, G. F. Assay of yeast mating reaction. Methods Enzymol. 194, 77–93 (1991).
Adams, A. E. & Pringle, J. R. Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 194, 729–731 (1991).
Pringle, J. R. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 194, 732–733 (1991).
James, P., Halladay, J. & Craig, E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 (1996).
Langle-Ronault, F. & Jacobs, E. Amethod for performing precise alterations in the yeast genome using a recyclable selectable marker. Nucleic Acids Res. 23, 3079–3081 (1995).
Acknowledgements
We thank Y. Anraku, R. Deschenes, G. Fink, E. Jacobs, P. James and P. Philippsen for strains and plasmids; M. Bassilana, M. Bienz, T. Bretscher, H. Pelham and S. Munro for comments and suggestions; and N. Lowe for assistance with sequencing. This work was supported by the Medical Research Council. A.N. was supported by fellowships from the Fonds der Chemischen Industrie and the EC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nern, A., Arkowitz, R. A GTP-exchange factor required for cell orientation. Nature 391, 195–198 (1998). https://doi.org/10.1038/34458
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/34458
This article is cited by
-
A framework for mapping, visualisation and automatic model creation of signal‐transduction networks
Molecular Systems Biology (2012)
-
Signaling mechanisms for regulation of chemotaxis
Cell Research (2005)
-
The ins and outs of cell-polarity decisions
Nature Cell Biology (2000)
-
Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating
Nature Cell Biology (2000)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.