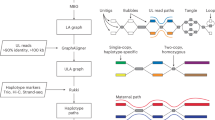
Abstract
Despite advances in long-read sequencing technologies, constructing a near telomere-to-telomere assembly is still computationally demanding. Here we present hifiasm (UL), an efficient de novo assembly algorithm combining multiple sequencing technologies to scale up population-wide near telomere-to-telomere assemblies. Applied to 22 human and two plant genomes, our algorithm produces better diploid assemblies at a cost of an order of magnitude lower than existing methods, and it also works with polyploid genomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
For the human reference genome, GRCh38 and CHM13v2; HiFi reads of HPRC year 2 samples except HG002: https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=submissions/1E2DD570-3B26-418B-B50F-5417F64C5679--HIFI_DEEPCONSENSUS/; ONT ultra-long reads of HPRC year 2 samples except HG002 (R9.4.1 flow cells): https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=submissions/90A1F283-2752-438B-917F-53AE76C9C43E--UCSC_HPRC_nanopore_Year2/; Hi-C reads of HPRC year 2 samples except HG002: https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=submissions/4C696EB9-9AD2-47A2-8011-2F43977CC4E0--Y2-HIC/; parental short reads of HPRC year 2 samples except HG002: https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=submissions/AD30A684-C7A8-4D24-89B2-040DFF021B0C--Y2_1000G_DATA/; HG002 HiFi reads (Google Cloud Storage): https://s3-us-west-2.amazonaws.com/human-pangenomics/NHGRI_UCSC_panel/HG002/hpp_HG002_NA24385_son_v1/PacBio_HiFi/20kb/m64011_190830_220126.Q20.fastq, https://s3-us-west-2.amazonaws.com/human-pangenomics/NHGRI_UCSC_panel/HG002/hpp_HG002_NA24385_son_v1/PacBio_HiFi/20kb/m64011_190901_095311.Q20.fastq, https://s3-us-west-2.amazonaws.com/human-pangenomics/NHGRI_UCSC_panel/HG002/hpp_HG002_NA24385_son_v1/PacBio_HiFi/15kb/m64012_190920_173625.Q20.fastq, https://s3-us-west-2.amazonaws.com/human-pangenomics/NHGRI_UCSC_panel/HG002/hpp_HG002_NA24385_son_v1/PacBio_HiFi/15kb/m64012_190921_234837.Q20.fastq; HG002 ultra-long reads (R9.4.1 flow cells, pass reads only): https://s3-us-west-2.amazonaws.com/human-pangenomics/NHGRI_UCSC_panel/HG002/nanopore/ultra-long/03_08_22_R941_HG002_1_Guppy_6.1.2_5mc_cg_prom_sup.tar, https://s3-us-west-2.amazonaws.com/human-pangenomics/NHGRI_UCSC_panel/HG002/nanopore/ultra-long/03_08_22_R941_HG002_2_Guppy_6.1.2_5mc_cg_prom_sup.tar, https://s3-us-west-2.amazonaws.com/human-pangenomics/NHGRI_UCSC_panel/HG002/nanopore/ultra-long/03_08_22_R941_HG002_3_Guppy_6.1.2_5mc_cg_prom_sup.tar; HG002 parental short reads: https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=working/HPRC_PLUS/HG002/raw_data/Illumina/parents/ and HG002 Hi-C reads: ‘HG002.HiC_1*’ from https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=working/HPRC_PLUS/HG002/raw_data/hic/downsampled/. All reads of HPRC year 1 samples (R9.4.1 flow cells for ONT reads): https://github.com/human-pangenomics/HPP_Year1_Data_Freeze_v1.0. All reads of Arabidopsis (R9.4.1 flow cells for ONT reads): https://ngdc.cncb.ac.cn/search/?dbId=gsa&q=CRA004538. All reads of potato (R9.4.1 flow cells for ONT ultra-long reads): https://ngdc.cncb.ac.cn/gsa/browse/CRA006012. Hifiasm (UL) assemblies of HPRC year 2 samples: ‘*hifiasm_v0.19.5*’ from https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=submissions/53FEE631-4264-4627-8FB6-09D7364F4D3B--ASM-COMP/. Verkko assemblies of HPRC year 2 samples: ‘*verkko_1.3.1*’ from https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=submissions/53FEE631-4264-4627-8FB6-09D7364F4D3B--ASM-COMP/. All evaluated HPRC year 1 and plant assemblies are available at https://zenodo.org/record/7996422 (ref. 25) and https://zenodo.org/record/7962930 (ref. 26), respectively.
Code availability
Hifiasm (UL) along with its source code is freely available at https://github.com/chhylp123/hifiasm.
References
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Wenger, A. M. et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37, 1155–1162 (2019).
Nurk, S. et al. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Res. 30, 1291–1305 (2020).
Kolmogorov, M., Yuan, J., Lin, Y. & Pevzner, P. A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019).
Luo, X., Kang, X. & Schönhuth, A. phasebook: haplotype-aware de novo assembly of diploid genomes from long reads. Genome Biol. 22, 299 (2021).
Porubsky, D. et al. Gaps and complex structurally variant loci in phased genome assemblies. Genome Res. 33, 496–510 (2023).
Jain, M. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Rautiainen, M. et al. Telomere-to-telomere assembly of diploid chromosomes with Verkko. Nat. Biotechnol. 41, 1474–1482 (2023).
Bankevich, A., Bzikadze, A. V., Kolmogorov, M., Antipov, D. & Pevzner, P. A. Multiplex de bruijn graphs enable genome assembly from long, high-fidelity reads. Nat. Biotechnol. 40, 1075–1081 (2022).
Rautiainen, M. & Marschall, T. MBG: minimizer-based sparse de Bruijn graph construction. Bioinformatics 37, 2476–2478 (2021).
Myers, E. W. The fragment assembly string graph. Bioinformatics 21, ii79–ii85 (2005).
Liao, W.-W. et al. A draft human pangenome reference. Nature 617, 312–324 (2023).
Lorig-Roach, R. et al. Phased nanopore assembly with Shasta and modular graph phasing with GFAse. Genome Res. https://genome.cshlp.org/content/early/2024/04/16/gr.278268.123 (2024).
Wang, B. et al. High-quality Arabidopsis thaliana genome assembly with nanopore and HiFi long reads. Genomics, Proteom. Bioinforma. 20, 4–13 (2022).
Naish, M. et al. The genetic and epigenetic landscape of the Arabidopsis centromeres. Science 374, eabi7489 (2021).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Bao, Z. et al. Genome architecture and tetrasomic inheritance of autotetraploid potato. Mol. Plant 15, 1211–1226 (2022).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Cheng, H. et al. Haplotype-resolved assembly of diploid genomes without parental data. Nat. Biotechnol. 40, 1332–1335 (2022).
Jain, C. Coverage-preserving sparsification of overlap graphs for long-read assembly. Bioinformatics 39, btad124 (2023).
Li, H., Feng, X. & Chu, C. The design and construction of reference pangenome graphs with minigraph. Genome Biol. 21, 265 (2020).
Edge, P., Bafna, V. & Bansal, V. HapCUT2: robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res. 27, 801–812 (2017).
Martin, M. et al. WhatsHap: fast and accurate read-based phasing. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/085050v2 (2016).
Cheng, H., Asri, M., Lucas, J., Koren, S. & Li, H. HPRC Y1 assemblies (HiFi + UL) evaluated in the hifiasm (UL) paper. Zenodo https://doi.org/10.5281/zenodo.7996421 (2023).
Cheng, H., Asri, M., Lucas, J., Koren, S. & Li, H. Plant assemblies evaluated in the hifiasm (UL) paper. Zenodo https://doi.org/10.5281/zenodo.7962929 (2023).
Acknowledgements
This study was supported by US National Institutes of Health (grant nos. R01HG010040, U01HG010971 and U41HG010972 to H.L. and K99HG012798 to H.C.). We thank the HPRC for making year 1 and year 2 datasets publicly available.
Author information
Authors and Affiliations
Contributions
H.C. and H.L. designed the algorithm, implemented hifiasm (UL) and drafted the paper. H.C. benchmarked hifiasm (UL) and other assemblers. M.A., J.L. and S.K. designed the evaluation of human genome assemblies.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Derek Bickhart and Antoine Limasset for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Lin Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
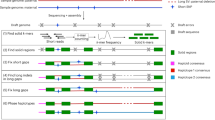
Extended Data Fig. 1 Accurate HiFi string graph combining PacBio HiFi and ONT ultra-long reads.
(a) Effect of contained reads in the string graph. Rectangles in orange and blue represent heterozygous HiFi reads from haplotype 1 and haplotype 2, respectively. Green rectangles are HiFi reads originating from homozygous regions, whereas red rectangles are contained reads. The string graph is constructed using all reads, except for two contained reads. (b) Hifiasm (UL) aligns ultra-long reads to the HiFi string graph with contained reads to alleviate the contained read problem. The alignment paths of ultra-long reads from haplotype 1 and haplotype 2 are represented by orange and blue lines, respectively. Despite being a contained read, h12 is retained as the critical read because it is covered by ultra-long reads u6 and u7. To ensure accurate graph cleaning, hifiasm (UL) also tracks the number of ultra-long reads that support each edge as its weight. For instance, the edge weight between h5 and h8 is 2 because ultra-long reads u4 and u5 cover it.
Supplementary information
Supplementary Information
Supplementary Tables 1–5, Fig. 1 and text.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cheng, H., Asri, M., Lucas, J. et al. Scalable telomere-to-telomere assembly for diploid and polyploid genomes with double graph. Nat Methods (2024). https://doi.org/10.1038/s41592-024-02269-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41592-024-02269-8