Abstract
Suicidal behavior is a complex disorder, with evidence for genetic risk independent of other genetic risk factors including psychiatric disorders. Since 1996, over 3000 DNA samples from Utah suicide decedents have been collected and banked for research use through the Utah Medical Examiner. In addition, over 12 000 Utah suicides were identified through examination of death certificates back to 1904. By linking this data with the Utah Population Database, we have identified multiple extended pedigrees with increased risk for suicide completion. A number of medical conditions co-occur with suicide, including asthma, and this study was undertaken to identify genetic risk common to asthma and suicide. This study tests the hypothesis that a particular comorbid condition may identify a more homogeneous genetic subgroup, facilitating the identification of specific genetic risk factors in that group. From pedigrees at increased risk for suicide, we identified three pedigrees also at significantly increased familial risk for asthma. Five suicide decedents from each of these pedigrees, plus an additional three decedents not from these pedigrees with diagnosed asthma, and 10 decedents with close relatives with asthma were genotyped. Results were compared with 183 publicly available unaffected control exomes from 1000 Genomes and CEPH (Centre d'etude du polymorphisme humain) samples genotyped on the same platform. A further 432 suicide decedents were also genotyped as non-asthma suicide controls. Genotyping was done using the Infinium HumanExome BeadChip. For analysis, we used the pedigree extension of Variant Annotation, Analysis and Search Tool (pVAAST) to calculate the disease burden of each gene. The Phenotype Driven Variant Ontological Re-ranking tool (Phevor) then re-ranked our pVAAST results in context of the phenotype. Using asthma as a seed phenotype, Phevor traversed biomedical ontologies and identified genes with similar biological properties to those known to result in asthma. Our top associated genes included those related to neurodevelopment or neural signaling (brain-derived neurotrophic factor (BDNF), neutral sphingomyelinase 2 (SMPD2), homeobox b2 (HOXB2), neural cell adhesion molecule (NCAM2), heterogeneous nuclear ribonucleoprotein A0 (HNRNPA0)), inflammation (free fatty acid receptor 2 (FFAR2)) and inflammation with additional evidence of neuronal involvement (oxidized low density lipoprotein receptor 1 (OLR1), toll-like receptor 3 (TLR3)). Of particular interest, BDNF has been previously implicated in both psychiatric disorders and asthma. Our results demonstrate the utility of combining pedigree and co-occurring phenotypes to identify rare variants associated with suicide risk in conjunction with specific co-occurring conditions.
Similar content being viewed by others
Introduction
In the United States, suicide is consistently in the top 10 leading causes of death, with over 38,000 reported in 2010.1 The Rocky Mountain states, in particular Utah, have elevated rates of completed suicide compared with the United States as a whole, with 17.5 and 11.8 suicides per 100 000, respectively, in 2009.2 Not only is the increased societal burden in Utah a compelling reason to conduct suicide research, but there are resources available to University of Utah researchers which make this an ideal setting. The Utah State Office of the Medical Examiner (OME) is centralized for the entire state and located on the University of Utah campus, which provides broad ascertainment, consistency in determining cause of death, and consistency in tissue/fluid collection for genetic and toxicology data. Information on suicide decedents from the OME is currently linked to pedigree, demographic and medical data available from the Utah Population Database (UPDB), an invaluable epidemiological resource with demographic, familial and medical data on 7.3 million individuals.3 This linking allows for identification of high-risk pedigrees, as well as characterization of psychiatric and physiological comorbidities. In conjunction with these resources, our laboratory recently reported the identification of several Utah pedigrees with increased risk for completed suicide.4
Several hypotheses have been proposed that attempt to explain the increased rates of suicide in Utah, specifically highlighting air quality and elevation.5 Utah, especially its large population centers, has some of the worst air quality in the United States.6 It has been shown that acute exposure to high levels of fine particulate matter and nitrogen dioxide increases the risk of suicide.7,8 Furthermore, Utah has the third highest mean elevation of the 50 states. Elevation has been correlated with increased suicide risk across the US and in South Korea.9, 10, 11 One possible explanation for this is increased cellular stress due to chronic hypoxia.11 Complementary to this idea are observations that pulmonary disease, especially asthma, increases risk for suicide.12 In a Swedish national cohort study, asthma increased the risk for suicide completion two-fold, after controlling for psychiatric disorders.13 Additional studies in the United States, Taiwan and South Korea have further established asthma as a risk factor for suicide and suicidal behavior.14, 15, 16, 17 Variation in the seasonality of suicide, with elevated rates during spring and autumn,18, 19, 20 could also partially be explained by increased exposure to seasonal airborne allergens,19,20 that have been shown to be triggers for both allergic rhinitis and asthma.21 At a clinical level, the association with asthma has come to the attention of the American Association of Suicidology, which has recommended regular evaluations for psychiatric conditions and suicidal ideation in patients with asthma (http://www.suicidology.org/resources/facts-statistics-current-research/current-research).
This study sought to identify genetic risk factors that may account for some of the observed comorbidity between asthma and suicide, with the future goal of improving interventions and treatment for suicidal behaviors, in this subgroup. As with many psychiatric disorders, the etiology of suicidal behavior is complex. Recent work has attempted to identify endophenotypes for suicidal behavior that better correspond to underlying risk factors.22,23 Along these lines, comorbid conditions like asthma could have a similar role, as segregating complex suicidal behavior into categories on the basis of comorbidity could reduce genetic heterogeneity and increase power to detect associations.
Materials and methods
Identification of suicide decedents and DNA collection through the OME, Utah State Department of Health
This study has been made possible through an ongoing collaboration between the University of Utah and the OME, with approval from each institutional review board. From December 1996 through July of 2011, we collected blood samples from 2215 Utah suicide decedents determined by the OME, as described previously.4 Blood samples were transported to the Translational Technologies and Resources (TTR) core laboratory, a component of the Center for Clinical and Translational Studies at the University of Utah. The TTR extracted DNA from the blood samples using a Qiagen AUTOPURE LS automated DNA extractor (Qiagen, Germantown, MD, USA) and DNA samples were stored at −80°C before genotyping.
Identification of pedigrees and suicide cases comorbid for asthma
Permission was obtained to link information from suicides determined by the OME where DNA had been collected to the Utah Population Database (UPDB).3 The UPDB contains information on over 7.3 million individuals, beginning with the 19th century Utah pioneers. The UPDB houses familial and demographic data from Vital Records made available through the State of Utah, including birth/death certificates, marriage/divorce licenses and driver licenses. The UPDB also contains medical information based on inpatient hospital discharge reports and ambulatory surgery records from outpatient providers, the majority within the state of Utah. In collaboration with biomedical researchers, the UPDB can link study subjects to familial data, identifying multi-generational pedigrees, and providing essential demographic and medical information, while maintaining privacy of pedigrees and individuals. The linking of suicide decedents with DNA from the OME to the UPDB for this study was conducted with additional permission from the Utah Resource for Genetic and Epidemiologic Research Committee.
Using records from 2215 suicide decedents from the OME with DNA, in addition to 12 850 records of other suicides ascertained from Utah death certificates going back to 1904, the UPDB identified pedigrees with increased familial risk for suicide as compared with the Utah population. To determine risk, the familial standardized incidence ratio (FSIR)24 was calculated for the founders of each pedigree by comparing the observed incidence of suicide in said pedigree with an expected incidence of suicide. The expected incidence of suicide was determined by generating a uniform distribution for suicide using multiple statewide pedigrees with similar age and sex compositions from the UPDB. The FSIR accounts for relatedness with the kinship coefficient, which measures the probability of an allele being identical by descent from a common ancestor. Simply put, the FSIR statistically measured the increased rate of suicide within each pedigree.
Within the pedigrees with significantly elevated FSIR for suicide (FSIR>2.0, P<0.05), we calculated the familial risk for asthma in a similar manner using electronic medical records linked to the UPDB to identify all pedigree members under each common founder couple with an asthma diagnosis and comparing pedigree incidence of asthma with matched pedigrees from the Utah population as described above. To identify asthma diagnoses, we used International Statistical Classification of Disease diagnostic codes (ICD-9, codes 493.##), which were ascertained from medical records available to the UPDB for all descendants of the identified common founders of the high-risk suicide pedigrees. This study focuses on suicides in three pedigrees at significantly increased risk for both suicide and asthma (n=15 pedigree suicide cases with DNA). In addition, we identified 10 other genotyped suicide cases who were not members of high-risk asthma/suicide pedigrees but had close relatives (1st or 2nd degree) diagnosed with asthma. Finally, the OME collected other phenotypic data, including current diagnosis of asthma and current medication, as part of the case report for each suicide decedent. These data were used to identify three additional genotyped suicide cases with asthma diagnoses not noted in the UPDB electronic records data who were not members of high-risk suicide pedigrees. Although these three diagnosed cases were missing from UPDB records, this simply suggests that these individuals were treated at medical facilities that do not currently share data with the UPDB. Our full suicide/asthma case cohort therefore included the 15 suicide cases in pedigrees at increased risk of both suicide and asthma, 10 suicides not in high-risk pedigrees but with a close family member with asthma, and three suicides not in high-risk pedigrees but with an asthma diagnosis (total n=28). All 28 subjects were White, dictating our selection of the race/ethnicity composition of comparison samples. All statistical comparisons of age and sex demographics between case cohort and the background Utah suicide cohort (not at comorbid risk for asthma) were conducted using R.25
Background control groups for molecular analyses
We generated a healthy control group on the basis of publicly available data from two sources. First, we used data from 90 CEPH Utah subjects with western European ancestry (CEU) who were genotyped by Illumina (San Diego, CA, USA) on the Infinium HumanExome Beadchip during chip development (www.support.illumina.com/array/array_kits/infinium_humanexome_beadchip_kit). These subjects are well matched to the Utah suicide cohort on demographic factors. The 90 subjects were composed of 30 trios, and the 30 child genotypes were discarded leaving 60 unrelated parents. Second, 135 subjects were selected from the 1000 Genomes Project, specifically CEPH Utah subjects (CEU) and British in England and Scotland (GBR) populations, and all had sequenced exomes. Alignment files were downloaded from 1000 Genomes (www.1000genomes.org) and Genome Analysis Toolkit (GATK v2.8)26 was used to compress the files using ReduceReads. CEU and GBR subjects were selected to match the ancestry of the suicide decedents.
Variants were called in GATK using Unified Genotyper, with a bed file used to force calls at each locus corresponding to variants on the HumanExome Chip. The output vcf file was converted to plink ped/map format using PLINK/SEQ (http://atgu.mgh.harvard.edu/plinkseq/).
Plink files from both sources were merged, and 12 subjects were identified as being duplications, having been both genotyped and sequenced. Concordance between genotyping and sequencing of these 12 duplicate subjects was 99.7%. For these duplications, only genotyping data was retained for further analysis, leaving 183 subjects in a combined control group.
Genotyping
A total of 472 suicide decedent subjects, including the suicide/asthma cohort were genotyped on the Infinium HumanExome Beadchip platform (v1.0, Illumina; http://genome.sph.umich.edu/wiki/Exome_Chip_Design), using protocols and standards outlined by Illumina. While features on this chip were taken from validated findings from sequencing studies, the platform is a straightforward genotyping chip. Samples were screened for quality and quantity before genotyping, and genotyping was conducted by the Microarray and Genomics Core Facility at the University of Utah Health Sciences Center. Five duplicated blinded samples showed 99.8% consistency between two separate batches.
Plink files were merged with background control files, and genotype data were screened in PLINK. We removed indels (140), variants missing in more than 10% of the total population (35 100) and uninformative homozygous variants (159 114). Twelve suicide decedents were removed for having call rates less than 75%. We retained 75 901 informative variants, with data from 460 suicide decedents and 183 healthy background controls. These final Plink format files were converted to individual files in GVF format (www.sequenceontology.org).
Genetic analysis
Genotype data were analyzed using a pedigree extension of the Variant Annotation, Analysis and Search Tool (VAAST v2.0.3).27, 28, 29 VAAST is a probabilistic disease-gene finder that uses variant and amino acid substitution frequency to calculate disease burden of each gene. The pedigree extension, pVAAST, incorporates extended pedigree relationships in the calculation of the composite likelihood ratio for each feature.29
We first annotated the variants for each subject using the VAAST Variant Analysis Tool with reference annotations available from www.sequenceontology.org/gff3.shtml, then used the VAAST Variant Selection Tool to generate a condensed file containing all annotated variants for all subjects. To separate cases from controls, we used Perl scripts distributed with VAAST. VAAST then compared the likelihood for each locus of a null model in which there is no difference in minor allele frequency between the ‘target’ samples and the primary comparison control samples, to an alternative model where the minor allele frequencies are different. VAAST uses a composite likelihood ratio, weighting variants by their potential for damaging effects according to amino acid substitution data. With a pedigree structure file, the pVAAST extension takes familial relationships into account in the composite likelihood ratio, prioritizing variants that tend to segregate with phenotype. Evidence across a gene is then aggregated to implicate particular genes. P-values are empirical, generated through a combination of a genome permutation process to account for the case–control allele frequency distribution, and a gene-drop simulation to account for pedigree structure.
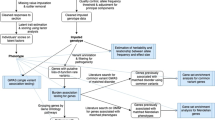
We conducted three pVAAST analyses (Figure 1). First, each variant was considered within the combined target group of suicide cases in pedigrees jointly significant for asthma and suicide plus singleton unrelated suicide cases with asthma or with a close relative with asthma (the asthma/suicide cohort). These cases were compared with the CEU/GBR background. P-values were determined with 1000 permutations. Genes were retained if P<0.05; 512 genes met this threshold. For the second analysis, again, the asthma/suicide cohort were compared with CEU/GBR background, but only variants contained in those 512 genes were tested, and P-values were computed with 109 permutations. The final pVAAST analysis compared these same 512 genes between the asthma/suicide cohort and the other Utah non-asthma suicides. These other non-asthma suicide cases are referred to as the background Utah suicide cohort for this analysis. One hundred and forty five genes met the threshold of P<0.05 in all three analyses and were included in the next step.
Shows a graphical depiction of the methods used for genetic analysis. The first pVAAST analysis used a low number of permutations across all genetic variants and compared the suicide/asthma cohort with the CEU/GBR background. Variants in the 512 genes meeting the P<0.05 threshold were used for pVAAST analyses 2 and 3. Analyses 2 and 3 used a high number of permutations across genetic variants in the 512 genes identified in analysis 1. Analysis 2 again compared the suicide/asthma cohort with the CEU/GBR background, and analysis 3 compared the suicide/asthma cohort with the non-asthma/Utah suicides background. The set of 145 genes meeting significance of P<0.05 in both analyses 2 and 3 were used as input for Phevor. Input P-values were masked as an agnostic approach, and Phevor was seeded with ‘asthma’ from three ontology databases. Twenty-one genes below P<0.01 were identified as candidate genes. CEU, western European ancestry; GBR, British in England and Scotland; Phevor, phenotype driven variant ontological re-ranking tool; pVAAST, pedigree extension of the Variant Annotation, Analysis and Search Tool.
The phenotype driven variant ontological re-ranking tool (Phevor)30 then re-ranked our pVAAST results in context of the phenotype. To approximate a more agnostic approach, we only used the gene names as input, masking the pVAAST generated P-values. Using ‘asthma’ as seed for the Human Phenotype Ontology, Disease Ontology and Pathway Ontology, Phevor traversed these biomedical ontologies and identified genes with similar biological properties to those known to result in the asthma phenotype.30 Individual variants in each gene with a Phevor P-value <0.01 were screened using PolyPhen231 and SIFT.32
Results
Identification of high-risk suicide pedigrees and high-risk asthma pedigrees
We identified 22 pedigrees with an excess of suicides (OME and death certificate), previously described in detail.4 Briefly, a first pass analysis of the UPDB identified 773 pedigrees with excess suicides (P⩽0.01). Of these, 183 pedigrees had five or more available DNA samples. A second pass of the top 30—ranked by each founder’s suicide FSIR—remaining pedigrees identified significant overlap between pedigree members of different pedigrees, and manual pruning resulted in 22 unique pedigrees. Three of these pedigrees were identified that had excess cases of asthma (P⩽0.0001, Table 1). Using Fisher’s exact test, there was no difference in the sex composition between each pedigree and the background Utah suicide cohort. The average age at death for the background Utah suicide cohort was 37.73 years. Pedigree 2 (two-sided t=−4.2, P=0.0006) had a significantly lower age at death (26.93 years) compared with this background suicide cohort, while pedigree 7a (t=−2.1, P=0.056) also trended in this direction (30.31 years). There was no age difference between pedigree 9 and the background suicide cohort. In addition, each pedigree has significantly elevated risk for other psychiatric disorders, including drug abuse (pedigrees 2, 7a, 9), alcohol use disorders (pedigrees 2 and 7a) and affective disorders (pedigree 2), shown in Table 1.
Identification of additional suicide cases at risk for asthma
Using data from the OME case reports and medical records available to the UPDB, we were able to identify additional decedents at risk for asthma who were not in the high-risk pedigrees, but who were also genotyped (Table 2). There were three decedents who had been diagnosed with asthma, three with at least one first-degree relative that had been diagnosed with asthma, and seven with at least one second-degree relative with an asthma diagnosis. There was no difference in sex composition between the group of at-risk individuals and the background Utah suicide cohort using Fisher’s exact test. There was no age difference between this group and the background cohort. Individuals in these groups had also been diagnosed with other psychiatric disorders, including depression, anxiety, drug abuse and alcohol use disorders, shown in Table 2.
Molecular analysis
Tables 3 and 4 show the top associated genes as determined by pVAAST and Phevor. Table 3 lists genes with a single variant ranked by pVAAST, whereas Table 4 lists genes with multiple variants ranked by pVAAST. Both tables list genes by Phevor ranking on the basis of Phevor score/P-value, with a P-value cutoff of 0.01. P-values from the two pVAAST analyses are reported. Both of these analyses used the asthma/suicide cohort as the target case group, but then compared against two backgrounds: (1) other UT suicides and (2) CEU/GBR from Illumina/1000 Genomes. In Table 3, genes were discarded if the variant only occurred in a single subject. Variant frequencies are reported for each group, using chromosome counts, as well as single-nucleotide polymorphism database frequencies. Table 3 lists PolyPhen2 and SIFT scores for each variant. Relevant phenotypic associations with these findings are presented in more detail in discussion section.
Discussion
This study expanded on previous research of the genetic etiology of suicide. Using data from suicide decedents from whom DNA was collected through July 2011 and additional suicide cases from death certificates going back to 1904, we used genealogical records in the Utah Population Database to identify 22 Utah pedigrees with at least twice the familial risk of suicide.4 Furthermore, linking medical records to members of these pedigrees allowed us to determine that three of these high-risk pedigrees were also at significantly elevated familial risk for asthma, although it remains possible that asthma cases were missing if diagnoses occurred out of state or if undiagnosed/untreated. Also worth noting are the unknown asthma and suicide rates in our 1000 genomes control group. There are extensive findings on the increased risk of suicide in individuals with asthma,12 but this is the first attempt at identifying genetic risk factors for suicide in individuals with asthma and with familial risk for asthma. We examined genetic variants in five individuals in each of the three high-risk suicide/asthma pedigrees, as well as thirteen additional individuals either diagnosed with asthma or with close relatives diagnosed with asthma.
Results from Infinium HumanExome Beadchip genotyping were first analyzed with pVAAST, which identified variants that segregated in target individuals compared with background. pVAAST scoring incorporates the deleteriousness of a particular variant, as well as familial relationships between subjects.27,28 Significant genes from pVAAST were then parsed with Phevor30 to identify which genes were more likely to be involved in asthma. As there are no ontologies directly related to suicide, we limited our Phevor search to asthma. Phevor allows for input of a list of candidate genes, which it expanded to all known interacting genes; therefore, as we continue to identify candidate genes for suicide, Phevor can be further implemented. It is important to note that this study design was limited to coding regions of the genome, as the HumanExome Beadchip primarily genotypes exomic variants. While regulatory variants in non-coding regions are not included in the current analysis, they will be explored in future analyses.
Brain-derived neurotrophic factor (BDNF) has been repeatedly implicated in both asthma33 and in psychiatric disease including suicide,34 but this is the first report of a more central role for BDNF in mediating risk for both asthma and suicide. Seven subjects, out of 28 in the asthma/suicide cohort, were heterozygous for the variant, including one subject in each of the three pedigrees, as well as one asthma case and three others with asthma relatives. Although the same variant was observed in each of the three independent pedigrees, there was no evidence in the UPDB that the three individuals shared a common ancestor. Although it has been hypothesized that the comorbidity observed between asthma and suicide could be mediated by variations in BDNF,35 this is the first study to have reported any findings. This particular variant in BDNF, rs66866077, predicted to be deleterious by both PolyPhen2 and SIFT, is only transcribed in mRNA transcript 6 (RefSeq: NM_170734.3), which translates to isoform c of precursor BDNF, proBDNF (RefSeq: NP_733930.1). proBDNF is itself a neurotrophin, binding p75 receptors, as well as being proteolytically cleaved into mature BDNF.36 The distinction and the interplay between the roles of proBDNF and mature BDNF have not been well established, but there is evidence of proBDNF involvement in early life neurodevelopment.37 Furthermore, the cleavage of proBDNF to mature BDNF may be attenuated by stress,38 which would corroborate observations of increased proBDNF in postmortem brains of suicide decedents.34 The results of this study suggest that specifically altering proBDNF may increase the risk of asthma and suicide. BDNF has been shown to be responsive to behavioral and pharmacological interventions like exercise39 and selective serotonin reuptake inhibitors.40,41 Ketamine, which shows promise as an acute treatment for depression42,43 and suicide behavior,42,44,45 may also depend on BDNF-mediated signaling.43,46 Another gene, SMPD2 (neutral sphingomyelinase 2) has been shown to facilitate oligodendrocyte cell death as a consequence of oxidative stress.47 Interestingly, SMPD2 also has a role in mediating neurotrophin signaling involving Trk receptors including NTRK2,48 which binds BDNF, and p75 receptors, which binds both BDNF and proBDNF.49 It is also worth noting that of the five asthma/suicide subjects with this variant in SMPD2, rs1048197, four of them also have the above BDNF variant.
We identified variants in several genes previously associated with inflammatory pathways. Although asthma is primarily an inflammatory disease,50 inflammation has been shown to have a role in psychiatric disease as well, including depression51 and bipolar disorder.52 The FFAR2 (free fatty acid receptor 2) may mediate the inflammatory response to short-chain free fatty acids generated by gut microbiota,53 although it is unclear whether FFAR2 has a direct role in respiratory inflammation.54 OLR1 (oxidized low density lipoprotein receptor 1) is induced by inflammatory cytokines and is upregulated in atherosclerosis.55 Variations in OLR1 have been associated with Alzheimer’s disease.56 TLR3 (Toll-like receptor 3) has an important role in the inflammatory response to viral infection,53 and has been implicated with asthma severity.57 TLR3 is also expressed in neurons,58 and expression has been shown to be increased in the prefrontal cortex of suicide decedents.59
Additional variants were identified in genes previously associated with the central nervous system. Homeobox B2 (HOXB2) has an important role in hindbrain development, specifically in oligodendrocyte sectioning.60 HNRNPA0 (heterogeneous nuclear ribonucleoprotein A0) was shown to be upregulated in the prefrontal cortex of schizophrenics.61 Neural cell adhesion molecule 2 (NCAM2) has a role in primary olfactory axon projections.62
Another variant that was consistently significant in the pVAAST analyses but was not identified by Phevor was rs34572680 in the purinergic receptor P2X subunit 3 (P2RX3). This non-synonymous variant (A71T, NP_002550.2) was deemed possibly damaging (PolyPhen2 score 0.853) and damaging (SIFT score 0.05). Eight of 28 subjects in the asthma/suicide cohort were heterozygous for this variant. The eight subjects include one subject from each pedigree, all the three asthma cases and two other subjects with close relatives with asthma. Although not previously suggested to be associated with asthma or suicide specifically, P2RX3 is involved in nociception,63 and variations in this gene have been associated with hyperalgesia.64 P2RX3 is expressed in the lung, among other tissues, and could respond to ATP released during hypoxic events.65 There is a well-established link between pain and suicide66 and the ongoing search for P2X3 antagonists may prove beneficial for the treatment of at-risk suicide patients.67,68
These results demonstrate the utility of combining pedigree and specific co-occurring phenotype information in the identification of rare variants associated with suicide risk. Through integrating pVAAST genetic analysis and Phevor phenotype association we have identified several genetic variants likely to increase risk for asthma and suicide, including a rare variant in BDNF. It is worth noting that although we lack power to identify epistatic variants, many of our top variants co-occurred in individual asthma/suicide decedents, for instance variants in SMPD2 and BDNF, two genes with evidence of functional relationships.48,49 We will continue to explore the possibility of epistatic variations, using Phevor to functionally cluster genes. Although the study would be more informative with a genotyped asthma/non-suicide control group, which could provide specificity as to whether variants confer risk for asthma/suicide/both, it is promising that our design identified BDNF, which has been linked extensively to both asthma33 and suicide.34 Future studies will also attempt to identify additional rare variants, as well as identify high-risk suicide pedigrees with other comorbid risk factors. There is also the possibility that these variants may interact with environmental exposure, that is, air pollution, which may further mediate risk for asthma and suicide. For instance, a variant in an asthma-related gene may be benign if the individual resides in an area with low air pollution exposure. When exposed to high air pollution, the normal genetic response may be compromised due to the variant, and downstream pathways may lead to increased asthma and suicide risk.
References
Centers for Disease Control and Prevention Leading causes of death. 2010. Available at www.cdc.gov/nchs/fastats/lcod.htm.
Centers for Disease Control and Prevention Utah Fact Sheet. Available at www.cdc.gov/nchs/pressroom/states/UT_2012.pdf.
Utah Population Database. Available at http://healthcare.utah.edu/huntsmancancerinstitute/research/updb/.
Coon HH, Darlington TM, Pimentel R, Smith KR, Huff CD, Hu H et al. Genetic risk factors in two Utah pedigrees at high risk for suicide. Transl Psychiatry 2013; 3: e325.
Haws CA, Gray DD, Yurgelun-Todd DA, Moskos M, Meyer LJ, Renshaw PF . The possible effect of altitude on regional variation in suicide rates. Med Hypotheses 2009; 73: 587–590.
American Lung Association State of the Air 2011. 2011. Available at http://www.lung.org/assets/documents/publications/state-of-the-air/state-of-the-air-2011-report.pdf.
Bakian A, Huber RS, Coon HH, Gray DD, McMahon WM, Renshaw PF . Acute air pollution exposure and risk of suicide completion. Am J Epidemiol 2014 (in press).
Kim C, Jung SH, Kang DR, Kim HC, Moon KT, Hur NW et al. Ambient particulate matter as a risk factor for suicide. Am J Psychiatry 2010; 167: 1100–1107.
Kim N, Mickelson JB, Brenner BE, Haws CA, Yurgelun-Todd DA, Renshaw PF . Altitude, gun ownership, rural areas, and suicide. Am J Psychiatry 2011; 168: 49–54.
Huber RS, Coon HH, Kim N, Renshaw PF, Kondo DG . Altitude is a risk factor for completed suicide in bipolar disorder. Med Hypotheses 2014; 82: 377–381.
Young SN . Elevated incidence of suicide in people living at altitude, smokers and patients with chronic obstructive pulmonary disease and asthma: possible role of hypoxia causing decreased serotonin synthesis. J Psychiatry Neurosci 2013; 38: 423–426.
Goodwin RD . Asthma and suicide: current knowledge and future directions. Curr Psychiatry Rep 2012; 14: 30–35.
Crump C, Sundquist K, Sundquist J, Winkleby MA . Sociodemographic, psychiatric and somatic risk factors for suicide: a Swedish national cohort study. Psychol Med 2013; 44: 279–289.
Goodwin RD, Demmer RT, Galea S, Lemeshow AR, Ortega AN, Beautrais A . Asthma and suicide behaviors: results from the Third National Health and Nutrition Examination Survey (NHANES III). J Psychiatr Res 2012; 46: 1002–1007.
Goodwin RD, Eaton WW . Asthma, suicidal ideation, and suicide attempts: findings from the Baltimore epidemiologic catchment area follow-up. Am J Public Health 2005; 95: 717–722.
Kuo C-J, VC-H Chen, Lee W-C, Chen WJ, Ferri CP, Stewart R et al. Asthma and suicide mortality in young people: a 12-year follow-up study. Am J Psychiatry 2010; 167: 1092–1099.
Chung SS, Joung KH . Risk factors related to suicidal ideation and attempted suicide: comparative study of Korean and American youth. J Sch Nurs 2012; 28: 448–458.
Woo J-M, Okusaga O, Postolache TT . Seasonality of suicidal behavior. Int J Environ Res Public Health 2012; 9: 531–547.
Postolache TT, Stiller JW, Herrell R, Goldstein Ma, Shreeram SS, Zebrak R et al. Tree pollen peaks are associated with increased nonviolent suicide in women. Mol Psychiatry 2005; 10: 232–235.
Messias EL, Clarke DE, Goodwin RD . Seasonal allergies and suicidality: results from the National Comorbidity Survey Replication. Acta Psychiatr Scand 2010; 122: 139–142.
Bousquet J, van Cauwenberge P, Khaltaev N . Allergic Rhinitis and Its Impact on Asthma. J Allergy Clin Immunol 2001; 108: S147–S334.
Mann JJ, Arango Va, Avenevoli S, Brent DA, Champagne FA, Clayton P et al. Candidate endophenotypes for genetic studies of suicidal behavior. Biol Psychiatry 2009; 65: 556–563.
Courtet P, Gottesman II, Jollant F, Gould TD . The neuroscience of suicidal behaviors: what can we expect from endophenotype strategies? Transl Psychiatry 2011; 1: e7.
Kerber RA . Method for calculating risk associated with family history of a disease. Genet Epidemiol 1995; 12: 291–301.
R Core Team. R: A language and environment for statistical computing, 2013.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303.
Yandell M, Huff CD, Hu H, Singleton M, Moore B, Xing J et al. A probabilistic disease-gene finder for personal genomes. Genome Res 2011; 21: 1529–1542.
Hu H, Huff CD, Moore B, Flygare S, Reese MG, Yandell M . VAAST 2.0: improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet Epidemiol 2013; 37: 622–634.
Hu H, Roach J, Coon HH, Guthery SL, Volkerding K, Margraf RL et al. A unified test of linkage analysis and rare-variant association. Nat Biotechnol 2014; 32: 663–669.
Singleton MV, Guthery SL, Voelkerding KV, Chen K, Kennedy B, Margraf RL et al. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am J Hum Genet 2014; 94: 599–610.
Adzhubei Ia, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249.
Kumar P, Henikoff S, Ng PC . Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081.
Prakash YS, Martin RJ . Brain-derived neurotrophic factor in the airways. Pharmacol Ther 2014; 143: 74–86.
Dwivedi Y . Brain-Derived Neurotrophic Factor and Suicide Pathogenesis. Ann Med 2010; 42: 87–96.
Ozek E, Ekici B . Asthma and suicide: possible role of brain-derived neurotrophic factor. Med Hypotheses 2011; 77: 261–262.
Barker PA . Whither proBDNF? Nat Neurosci 2009; 12: 105–106.
Wong J, Webster MJ, Cassano H, Weickert CS . Changes in alternative brain-derived neurotrophic factor transcript expression in the developing human prefrontal cortex. Eur J Neurosci 2009; 29: 1311–1322.
Tognoli C, Rossi F, Di Cola F, Baj G, Tongiorgi E, Terova G et al. Acute stress alters transcript expression pattern and reduces processing of proBDNF to mature BDNF in Dicentrarchus labrax. BMC Neurosci 2010; 11: 4.
Zoladz JA, Pilc A . The effect of physical activity on the brain derived neurotrophic factor: from animal to human studies. J Physiol Pharmacol 2010; 61: 533–541.
Duman RS, Monteggia LM . A neurotrophic model for stress-related mood disorders. Biol Psychiatry 2006; 59: 1116–1127.
Russo-Neustadt A, Beard RC, Cotman CW . Exercise, antidepressant medications, and enhanced brain derived neurotrophic factor expression. Neuropsychopharmacology 1999; 21: 679–682.
Howland RH . Ketamine for the treatment of depression. J Psychosoc Nurs Ment Health Serv 2013; 51: 11–14.
Akinfiresoye L, Tizabi Y . Antidepressant effects of AMPA and ketamine combination: role of hippocampal BDNF, synapsin, and mTOR. Psychopharmacology (Berl) 2013; 230: 291–298.
Price RB, Iosifescu DV, Murrough JW, Chang LC, Al Jurdi RK, Iqbal SZ et al. Effects of ketamine on explicit and implicit suicidal cognition: a randomized controlled trial in treatment-resistant depression. Depress Anxiety 2014; 343: 335–343.
De Gioannis A, De Leo D . Oral ketamine augmentation for chronic suicidality in treatment-resistant depression. Aust N Z J Psychiatry 2014; 48: 686.
Haile CN, Murrough JW, Iosifescu DV, Chang LC, Al Jurdi RK, Foulkes AL et al. Plasma brain derived neurotrophic factor (BDNF) and response to ketamine in treatment-resistant depression. Int J Neuropsychopharmacol 2014; 17: 331–336.
Jana A, Pahan K . Oxidative stress kills human primary oligodendrocytes via neutral sphingomyelinase: implications for multiple sclerosis. J Neuroimmune Pharmacol 2007; 2: 184–193.
Candalija A, Cubí R, Ortega A, Aguilera J, Gil C . Trk receptors need neutral sphingomyelinase activity to promote cell viability. FEBS Lett 2014; 588: 167–174.
Bilderback TR, Gazula VR, Dobrowsky RT . Phosphoinositide 3-kinase regulates crosstalk between Trk A tyrosine kinase and p75(NTR)-dependent sphingolipid signaling pathways. J Neurochem 2001; 76: 1540–1551.
Lemanske RF, Busse WW . Asthma: clinical expression and molecular mechanisms. J Allergy Clin Immunol 2010; 125: S95–102.
Rosenblat JD, Cha DS, Mansur RB, McIntyre RS . Inflamed moods: A review of the interactions between inflammation and mood disorders. Prog Neuropsychopharmacol Biol Psychiatry 2014; 53: 23–34.
Stertz L, Magalhães PVS, Kapczinski F . Is bipolar disorder an inflammatory condition? The relevance of microglial activation. Curr Opin Psychiatry 2013; 26: 19–26.
Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009; 461: 1282–1286.
Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014; 20: 159–166.
Pirillo A, Norata GD, Catapano AL . LOX-1, OxLDL, and atherosclerosis. Mediators Inflamm 2013; 2013: 152786.
D’Introno A, Solfrizzi V, Colacicco AM, Capurso C, Torres F, Capurso SA et al. Polymorphisms in the oxidized low-density lipoprotein receptor-1 gene and risk of Alzheimer’s disease. J Gerontol A Biol Sci Med Sci 2005; 60: 280–284.
Matsukura S, Kurokawa M, Homma T, Watanabe S, Suzuki S, Ieki K et al. Basic research on virus-induced asthma exacerbation: inhibition of inflammatory chemokine expression by fluticasone propionate. Int Arch Allergy Immunol 2013; 161: 84–92.
Lafon M, Megret F, Lafage M, Prehaud C . The innate immune facet of brain: human neurons express TLR-3 and sense viral dsRNA. J Mol Neurosci 2006; 29: 185–194.
Pandey GN, Rizavi HS, Ren X, Bhaumik R, Dwivedi Y . Toll-like receptors in the depressed and suicide brain. J Psychiatr Res 2014; 53: 62–68.
Miguez A, Ducret S, Di Meglio T, Parras C, Hmidan H, Haton C et al. Opposing roles for Hoxa2 and Hoxb2 in hindbrain oligodendrocyte patterning. J Neurosci 2012; 32: 17172–17185.
Martins-de-Souza D, Gattaz WF, Schmitt A, Rewerts C, Maccarrone G, Dias-Neto E et al. Prefrontal cortex shotgun proteome analysis reveals altered calcium homeostasis and immune system imbalance in schizophrenia. Eur Arch Psychiatry Clin Neurosci 2009; 259: 151–163.
Winther M, Berezin V, Walmod PS . NCAM2/OCAM/RNCAM: cell adhesion molecule with a role in neuronal compartmentalization. Int J Biochem Cell Biol 2012; 44: 441–446.
Donnelly-Roberts D, McGaraughty S, Shieh C, Honore P, Jarvis MF . Painful purinergic receptors. J Pharmacol Exp Ther 2008; 324: 409–415.
Giniatullin R, Nistri A, Fabbretti E . Molecular mechanisms of sensitization of pain-transducing P2X3 receptors by the migraine mediators CGRP and NGF. Mol Neurobiol 2008; 37: 83–90.
Burnstock G . Purinergic mechanosensory transduction and visceral pain. Mol Pain 2009; 5: 69.
Elman I, Borsook D, Volkow ND . Pain and suicidality: insights from reward and addiction neuroscience. Prog Neurobiol 2013; 109: 1–27.
Stahl SM . Mechanism of action of ketamine. CNS Spectr 2013; 18: 171–174.
Ford AP . In pursuit of P2X3 antagonists: novel therapeutics for chronic pain and afferent sensitization. Purinergic Signal 2012; 8: 3–26.
Acknowledgements
MHR01099134, VISN 19 MIRECC, USTAR, GCRCM01RR025764, AGR01022095, Clark Tanner Foundation, Sharon Kae Lehr Endowed Research Fund, Odyssey Program. Partial support for data within the Utah Population Database (UPDB) was provided by the University of Utah Huntsman Cancer Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Darlington, T., Pimentel, R., Smith, K. et al. Identifying rare variants for genetic risk through a combined pedigree and phenotype approach: application to suicide and asthma. Transl Psychiatry 4, e471 (2014). https://doi.org/10.1038/tp.2014.111
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2014.111
This article is cited by
-
Antidepressants normalize elevated Toll-like receptor profile in major depressive disorder
Psychopharmacology (2016)
-
Understanding and predicting suicidality using a combined genomic and clinical risk assessment approach
Molecular Psychiatry (2015)