Featured
-
-
Perspective |
 Hold out the genome: a roadmap to solving the cis-regulatory code
Hold out the genome: a roadmap to solving the cis-regulatory code
A roadmap towards solving the cis-regulatory code using a combination of machine learning and massively parallel assays of exogenous DNA is proposed.
- Carl G. de Boer
- & Jussi Taipale
-
Article |
 Transfer learning enables predictions in network biology
Transfer learning enables predictions in network biology
A context-aware, attention-based deep learning model pretrained on single-cell transcriptomes enables predictions in settings with limited data in network biology and could accelerate discovery of key network regulators and candidate therapeutic targets.
- Christina V. Theodoris
- , Ling Xiao
- & Patrick T. Ellinor
-
Article
| Open Access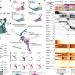 Inferring and perturbing cell fate regulomes in human brain organoids
Inferring and perturbing cell fate regulomes in human brain organoids
A multi-omic atlas of brain organoid development facilitates the inference of an underlying gene regulatory network using the newly developed Pando framework and shows—in conjunction with perturbation experiments—that GLI3 controls forebrain fate establishment through interaction with HES4/5 regulomes.
- Jonas Simon Fleck
- , Sophie Martina Johanna Jansen
- & Barbara Treutlein
-
Article
| Open Access Cell-type specialization is encoded by specific chromatin topologies
Cell-type specialization is encoded by specific chromatin topologies
A new technique called immunoGAM, which combines genome architecture mapping (GAM) with immunoselection, enabled the discovery of specialized chromatin conformations linked to gene expression in specific cell populations from mouse brain tissues.
- Warren Winick-Ng
- , Alexander Kukalev
- & Ana Pombo
-
Article |
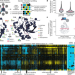 Systems-level effects of allosteric perturbations to a model molecular switch
Systems-level effects of allosteric perturbations to a model molecular switch
Interface mutations in the GTPase switch protein Gsp1 (the yeast homologue of human RAN) allosterically affect the kinetics of the switch cycle, revealing a systems-level mechanism of multi-specificity.
- Tina Perica
- , Christopher J. P. Mathy
- & Tanja Kortemme
-
Article |
 MIR-NATs repress MAPT translation and aid proteostasis in neurodegeneration
MIR-NATs repress MAPT translation and aid proteostasis in neurodegeneration
The natural antisense transcript MAPT-AS1 interferes with translation of mRNA transcript into tau protein in the brain and may represent a general mechanism for controlling levels of intrinsically disordered proteins, with particular relevance for neurodegeneration.
- Roberto Simone
- , Faiza Javad
- & Rohan de Silva
-
Article |
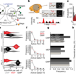 Single-molecule imaging of transcription dynamics in somatic stem cells
Single-molecule imaging of transcription dynamics in somatic stem cells
Single-molecule fluorescence in situ hybridization and live-cell imaging are used to study the contribution of transcriptional noise to stem cell heterogeneity, revealing that stochastic transcription dynamics are conducive to concomitant stem-cell maintenance and tissue homeostasis.
- Justin C. Wheat
- , Yehonatan Sella
- & Ulrich Steidl
-
Letter |
 An extracellular network of Arabidopsis leucine-rich repeat receptor kinases
An extracellular network of Arabidopsis leucine-rich repeat receptor kinases
A high-throughput assay is used to analyse 40,000 potential extracellular domain interactions of a large family of plant cell surface receptors (LRR-RKs) and provide a cell surface interaction network for these receptors.
- Elwira Smakowska-Luzan
- , G. Adam Mott
- & Youssef Belkhadir
-
Letter |
 A global resource allocation strategy governs growth transition kinetics of Escherichia coli
A global resource allocation strategy governs growth transition kinetics of Escherichia coli
A new approach to modelling bacterial growth removes the need to know kinetic parameters for metabolic and regulatory processes and can be used to model adaptive processes such as antibiotic responses and ecological dynamics.
- David W. Erickson
- , Severin J. Schink
- & Terence Hwa
-
Letter |
 Intersecting transcription networks constrain gene regulatory evolution
Intersecting transcription networks constrain gene regulatory evolution
Epistatic interactions, whereby a mutation's effect is contingent on another mutation, have been shown to constrain evolution within single proteins, and how such interactions arise in gene regulatory networks has remained unclear; here the appearance of pheromone-response regulator binding sites in the regulatory DNA of the a-specific genes of Saccharomyces cerevisiae are shown to have required specific changes in a second pathway during the evolution from its common ancestor with Candida albicans.
- Trevor R. Sorrells
- , Lauren N. Booth
- & Alexander D. Johnson
-
Letter |
 Condensin-driven remodelling of X chromosome topology during dosage compensation
Condensin-driven remodelling of X chromosome topology during dosage compensation
Genome-wide chromosome conformation capture analysis in C. elegans reveals that the dosage compensation complex, a condensin complex, remodels the X chromosomes of hermaphrodites into a sex-specific topology distinct from autosomes while regulating gene expression chromosome-wide.
- Emily Crane
- , Qian Bian
- & Barbara J. Meyer
-
Letter |
 hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1
hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1
A method, termed hiCLIP, has been developed to determine the RNA duplexes bound by RNA-binding proteins, revealing an unforeseen prevalence of long-range duplexes in 3′ untranslated regions (UTRs), and a decreased incidence of SNPs in duplex-forming regions; the results also show that RNA structure is able to regulate gene expression.
- Yoichiro Sugimoto
- , Alessandra Vigilante
- & Jernej Ule
-
Article
| Open Access Integrative analysis of 111 reference human epigenomes
Integrative analysis of 111 reference human epigenomes
This study describes the integrative analysis of 111 reference human epigenomes, profiled for histone modification patterns, DNA accessibility, DNA methylation and RNA expression; the results annotate candidate regulatory elements in diverse tissues and cell types, their candidate regulators, and the set of human traits for which they show genetic variant enrichment, providing a resource for interpreting the molecular basis of human disease.
- Anshul Kundaje
- , Wouter Meuleman
- & Manolis Kellis
-
Article |
 An Arabidopsis gene regulatory network for secondary cell wall synthesis
An Arabidopsis gene regulatory network for secondary cell wall synthesis
The full complement of transcriptional regulators that affect synthesis of the plant secondary cell wall remains largely undetermined; here, the network of protein–DNA interactions controlling secondary cell wall synthesis of Arabidopsis thaliana is determined, showing that gene expression is regulated by a series of feed-forward loops to ensure that the secondary cell wall is deposited at the right time and in the right place.
- M. Taylor-Teeples
- , L. Lin
- & S. M. Brady
-
Article |
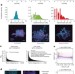 Deconstructing transcriptional heterogeneity in pluripotent stem cells
Deconstructing transcriptional heterogeneity in pluripotent stem cells
This study uses single-cell expression profiling of pluripotent stem cells after various perturbations, and uncovers a high degree of variability that can be inherited through cell divisions—modulating microRNA or external signalling pathways induces a ground state with reduced gene expression heterogeneity and a distinct chromatin profile.
- Roshan M. Kumar
- , Patrick Cahan
- & James J. Collins
-
Article
| Open Access Architecture of the human regulatory network derived from ENCODE data
Architecture of the human regulatory network derived from ENCODE data
A description is given of the ENCODE consortium’s efforts to examine the principles of human transcriptional regulatory networks; the results are integrated with other genomic information to form a hierarchical meta-network where different levels have distinct properties.
- Mark B. Gerstein
- , Anshul Kundaje
- & Michael Snyder

 Transcription–replication interactions reveal bacterial genome regulation
Transcription–replication interactions reveal bacterial genome regulation