Abstract
The plant cell wall is an important factor for determining cell shape, function and response to the environment. Secondary cell walls, such as those found in xylem, are composed of cellulose, hemicelluloses and lignin and account for the bulk of plant biomass. The coordination between transcriptional regulation of synthesis for each polymer is complex and vital to cell function. A regulatory hierarchy of developmental switches has been proposed, although the full complement of regulators remains unknown. Here we present a protein–DNA network between Arabidopsis thaliana transcription factors and secondary cell wall metabolic genes with gene expression regulated by a series of feed-forward loops. This model allowed us to develop and validate new hypotheses about secondary wall gene regulation under abiotic stress. Distinct stresses are able to perturb targeted genes to potentially promote functional adaptation. These interactions will serve as a foundation for understanding the regulation of a complex, integral plant component.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Brown, D. M., Zeef, L. A. H., Ellis, J., Goodacre, R. & Turner, S. R. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295 (2005)
Persson, S., Wei, H., Milne, J., Page, G. P. & Somerville, C. R. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl Acad. Sci. USA 102, 8633–8638 (2005)
Carlsbecker, A. et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321 (2010)
Mitsuda, N., Seki, M., Shinozaki, K. & Ohme-Takagi, M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006 (2005)
Brady, S. M. et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 (2007)
Zhong, R., Lee, C., Zhou, J., McCarthy, R. L. & Ye, Z.-H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782 (2008)
Gaudinier, A. et al. Enhanced Y1H assays for Arabidopsis. Nature Methods 8, 1053–1055 (2011)
Kim, W.-C., Ko, J.-H. & Han, K.-H. Identification of a cis-acting regulatory motif recognized by MYB46, a master transcriptional regulator of secondary wall biosynthesis. Plant Mol. Biol. 78, 489–501 (2012)
Yamaguchi, M. et al. VASCULAR-RELATED NAC-DOMAIN 7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 66, 579–590 (2011)
Zhou, J., Lee, C., Zhong, R. & Ye, Z. H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248–266 (2009)
Hussey, S. G., Mizrachi, E., Creux, N. M. & Myburg, A. A. Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front. Plant Sci. 4, 325 (2013)
Walhout, A. J. M. What does biologically meaningful mean? A perspective on gene regulatory network validation. Genome Biol. 12, 109 (2011)
Kim, W.-C., Kim, J.-Y., Ko, J.-H., Kang, H. & Han, K.-H. Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol. Biol. 85, 589–599 (2014)
Ahnert, S. E. Power graph compression reveals dominant relationships in genetic transcription networks. Mol. Biosyst. 9, 2681–2685 (2013)
Kim, W.-C. et al. MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J. 73, 26–36 (2013)
del Pozo, J. C., Diaz-Trivino, S., Cisneros, N. & Gutierrez, C. The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18, 2224–2235 (2006)
del Pozo, J. C., Boniotti, M. B. & Gutierrez, C. Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCFAtSKP2 pathway in response to light. Plant Cell 14, 3057–3071 (2002)
de Jager, S. M., Menges, M., Bauer, U. M. & Murray, J. A. H. Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Mol. Biol. 47, 555–568 (2001)
Mariconti, L. et al. The E2F family of transcription factors from Arabidopsis thaliana: novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 277, 9911–9919 (2002)
Kosugi, S. & Ohashi, Y. Interaction of the Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol. 128, 833–843 (2002)
de Jager, S. et al. Dissecting regulatory pathways of G1/S control in Arabidopsis: common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol. Biol. 71, 345–365 (2009)
Heckmann, S. et al. The E2F transcription factor family regulates CENH3 expression in Arabidopsis thaliana. Plant J. 68, 646–656 (2011)
del Pozo, J. C., Diaz-Trivino, S., Cisneros, N. & Gutierrez, C. The E2FC-DPB transcription factor controls cell division, endoreplication and lateral root formation in a SCFSKP2A-dependent manner. Plant Signal. Behav. 2, 273–274 (2007)
Yamaguchi, M. et al. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22, 1249–1263 (2010)
Yamaguchi, M. et al. VASCULAR-RELATED NAC-DOMAIN6 (VND6) and VND7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 153, 906–914 (2010)
Wenkel, S., Emery, J., Hou, B.-H., Evans, M. M. S. & Barton, M. K. A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 19, 3379–3390 (2007)
Zhong, R. & Ye, Z.-H. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 53, 368–380 (2012)
Ohashi-Ito, K., Oda, Y. & Fukuda, H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473 (2010)
Pyo, H., Demura, T. & Fukuda, H. TERE; a novel cis-element responsible for a coordinated expression of genes related to programmed cell death and secondary wall formation during differentiation of tracheary elements. Plant J. 51, 955–965 (2007)
Dinneny, J. R. et al. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320, 942–945 (2008)
Iyer-Pascuzzi, A. S. et al. Cell identity regulators link development and stress responses in the Arabidopsis root. Dev. Cell 21, 770–782 (2011)
Gifford, M. L., Dean, A., Gutierrez, R. A., Coruzzi, G. M. & Birnbaum, K. D. Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl Acad. Sci. USA 105, 803–808 (2008)
Brady, S. M. et al. A stele-enriched gene regulatory network in the Arabidopsis root. Mol. Syst. Biol. 7, 459 (2011)
Gong, W. et al. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 135, 773–782 (2004)
Paz-Ares, J. & The REGIA Consortium. REGIA, an EU project on functional genomics of transcription factors from Arabidopsis thaliana. Comp. Funct. Genomics 3, 102–108 (2002)
Underwood, B. A., Vanderhaeghen, R., Whitford, R., Town, C. D. & Hilson, P. Simultaneous high-throughput recombinational cloning of open reading frames in closed and open configurations. Plant Biotechnol. J. 4, 317–324 (2006)
Yamada, K., Lim, J., Dale, J. & Chen, H. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302, 842–846 (2003)
Pruneda-Paz, J. L., Breton, G., Para, A. & Kay, S. A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485 (2009)
Kubo, M. et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860 (2005)
Hawker, N. P. & Bowman, J. L. Roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 135, 2261–2270 (2004)
Updegraff, D. Semimicro determination of cellulose in biological materials. Anal. Biochem. 32, 420–424 (1969)
Scott, T. A. & Melvin, E. H. Determination of dextran with anthrone. Anal. Chem. 25, 1656–1661 (1953)
Liu, Y., Burch-Smith, T., Schiff, M., Feng, S. & Dinesh-Kumar, S. P. Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 279, 2101–2108 (2004)
Voinnet, O., Rivas, S., Mestre, P. & Baulcombe, D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956 (2003)
Walley, J. W. et al. Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. 3, e172 (2007)
Nusinow, D. A. et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402 (2011)
Pearson, R. D. et al. puma: a Bioconductor package for propagating uncertainty in microarray analysis. BMC Bioinformatics 10, 211 (2009)
Mordelet, F. & Vert, J.-P. SIRENE: supervised inference of regulatory networks. Bioinformatics 24, i76–i82 (2008)
Huynh-Thu, V. A., Irrthum, A., Wehenkel, L. & Geurts, P. Inferring regulatory networks from expression data using tree-based methods. PLoS ONE 5, e12776 (2010)
Greenfield, A., Madar, A., Ostrer, H. & Bonneau, R. DREAM4: Combining genetic and dynamic information to identify biological networks and dynamical models. PLoS ONE 5, e13397 (2010)
Haury, A.-C., Mordelet, F., Vera-Licona, P. & Vert, J.-P. TIGRESS: Trustful Inference of Gene REgulation using Stability Selection. BMC Syst. Biol. 6, 145 (2012)
Küffner, R., Petri, T., Tavakkolkhah, P., Windhager, L. & Zimmer, R. Inferring gene regulatory networks by ANOVA. Bioinformatics 28, 1376–1382 (2012)
Marbach, D. et al. Wisdom of crowds for robust gene network inference. Nature Methods 9, 796–804 (2012)
Acknowledgements
We thank M. Tierney (University of Vermont) for 35S::GFP seeds, T. Demura for VND7 resources, M.K. Barton for REV:GR seeds, E.P. Spalding for advice on manuscript revision, and C. Gutierrez for E2Fc RNAi and E2Fc N-terminal deletion overexpressor seeds and useful discussion. This research was supported by the Office of Science (BER) Department of Energy Grant DE-FG02-08ER64700DE (to S.P.H. and S.A.K.), National Institute of General Medical Sciences of the National Institutes of Health under award numbers RO1GM056006 and RC2GM092412 (to S.A.K.), National Institute of Health (R01GM107311) and National Science Foundation (IOS-1036491 and IOS-1352478) to K.D., USDA CRIS 1907-21000-030 to D.W. and L.F., a Royal Society UK Fellowship (to S.E.A.), and UC Davis Startup Funds and a Hellman Fellowship (to S.M.B).
Author information
Authors and Affiliations
Contributions
M.T.-T., L.L. and M.d.L. contributed equally to this work. T.W.T. and A.G. contributed equally to this work. S.M.B. and S.P.H. contributed equally to this work. M.T.-T., L.L., M.d.L., S.M.B., and S.P.H. designed the research. M.T.-T., L.L., M.d.L., A.G., G.X., N.F.Y., G.M.T., M.T.V., R.L., P.P.H., C.W., and K.D. performed the research. M.T.-T., L.L., G.T., T.W.T., N.T., J.C., M.P., D.K., I.T., S.E.A., S.M.B. and S.P.H. analysed the data. L.Z., D.W., G.B., J.L.P.-P., and S.A.K. contributed new reagents/analytic tools. M.T.-T., L.L., G.M.T., S.M.B. and S.P.H. wrote the article. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
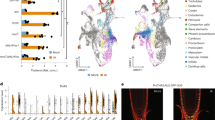
Extended Data Figure 1 Number of novel and previously described protein–DNA interactions and transcription factors involved in secondary cell wall biosynthesis and xylem development.
a, b, Venn diagrams of overlap between previously reported19 interactions (a) or transcription factors (b) and those of the xylem-specific gene regulatory network. *Includes genes that were not included in the yeast one hybrid screen.
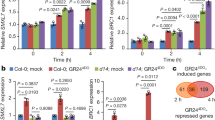
Extended Data Figure 2 Activation or repression of VND7 by E2Fc is dynamic and dose-dependent.
a, Intensity of LUC bioluminescence quantified using Andor Solis image analysis software. Data are means ± s.d. (n = 20). Asterisks denote significance at P < 0.05 determined by Student’s t-test. b, Quantitative PCR with reverse transcription of E2Fc and VND7 transcripts in ΔN-E2Fc (E2Fc overexpressor line lacking the N-terminal domain) expressing plants versus Col-0 control. Red dashed line marks the point at which VND7 is unchanged compared to control. Each data point is an individual biological replicate with 3 technical replicates. c, 3-week-old tobacco leaves were infiltrated with the p19 silencing inhibitor and either the reporter VND7p::GUS or VND7p::GUS and either 35S::E2Fc::MYC or 35S::RBR::GFP, or both. Extracted protein was then used in a quantitative MUG fluorescent assay, where relative fluorescence was measured 60 min after incubation with substrate. Data are means ± s.d., n = 3.
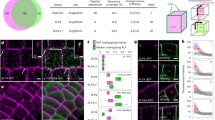
Extended Data Figure 3 Binding of NST2 and SND1 to fragments of CESA7, CESA8, and KOR promoters.
a–f, Electrophoretic mobility shift assays showing NST2 (a–d) and SND1 (e–f) protein specifically binds the promoters of cellulose-associated genes. Probe was incubated in the absence or presence of GST or GST:SND1 protein extracts. The arrowheads indicate the specific protein–DNA complexes, while arrows indicate free probe.
Extended Data Figure 4 Sub-networks of network genes differentially expressed in response to iron deprivation of high salinity.
a, b, Sub-network of genes with q values of ≤ 0.01 and whose fold change between mean expression values was ≥ 1.5 in either direction in iron deprivation (a) or high NaCl (b) stress microarray data set. Nodes are coloured according to in-degree as shown on scale bars below sub-networks. Transcription factors with the highest in-degree are labelled and indicated with a black circle.
Extended Data Figure 5 The reconstructed gene regulatory consensus network based on analysis of the iron-deprivation expression data set by different network inference methods.
a, Unsupervised; b, supervised in the first pass; c, supervised after the validated two connections have been added in the training set. Edge transparency denotes P ≤ 0.06 for the Pearson correlation coefficient (PCC); edge width is proportional to PCC; edge value corresponds to the total edge score; a greater value corresponds to a more significant score. Yellow and red nodes correspond to transcription factor and target gene nodes, respectively; black and blue edges denote Y1H-derived and inferred interactions, respectively.
Extended Data Figure 6 Iron deprivation and NaCl stress influences lignin and phenylpropanoid biosynthesis associated gene expression.
a, No change was observed in the expression of 4CL1::GFP in 4 days after imbibition (DAI) roots transferred to a control media (left, n = 4) or media with 140 mM NaCl for 48 h (right, n = 4). b, Increased fuchsin staining of xylem cells as well as of cell walls of non-vascular cells in 4 DAI roots transferred to a control media (left) or media with an iron chelator for 72 h (right). c, No change was observed in the expression of VND7::YFP in 4 DAI roots transferred to a control media (left, n = 4) or media with an iron chelator for 72 h (right, n = 5).
Extended Data Figure 7 The reconstructed gene regulatory consensus network based on analysis of the salt-stress expression data set by different network inference methods.
a, Unsupervised; b, supervised in the first pass; c, supervised after the validated two connections have been added in the training set. Edge transparency denotes P ≤ 0.06 for the Pearson correlation coefficient (PCC); edge width is proportional to PCC; edge value corresponds to the total edge score; a greater value corresponds to a more significant score. Yellow and red nodes correspond to transcription factor and target gene nodes, respectively; black and blue edges denote Y1H-derived and inferred interactions, respectively.
Extended Data Figure 8 Schematic diagram of dual-luciferase reporter vector development.
a, Three distinct donor vectors harbouring either the transcription factor, VP64 activation domain fused to the 35S minimal promoter, or a promoter fragment. b, The dual reporter vector, pLAH-LARm, is then recombined with the three donor vectors to generate the single reporter vector (c).
Supplementary information
Supplementary information
This zipped file contains Supplementary Tables 1-10 and a Supplementary Guide containing full table legends. (ZIP 424 kb)
Rights and permissions
About this article
Cite this article
Taylor-Teeples, M., Lin, L., de Lucas, M. et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575 (2015). https://doi.org/10.1038/nature14099
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14099
This article is cited by
-
Basic helix-loop-helix (bHLH) gene family in rye (Secale cereale L.): genome-wide identification, phylogeny, evolutionary expansion and expression analyses
BMC Genomics (2024)
-
Combined metabolome and transcriptome analysis reveals a critical role of lignin biosynthesis and lignification in stem-like pneumatophore development of the mangrove Avicennia marina
Planta (2024)
-
GhHB14_D10 and GhREV_D5, two HD-ZIP III transcription factors, play a regulatory role in cotton fiber secondary cell wall biosynthesis
Plant Cell Reports (2024)
-
Characterization of ZmCesAs for Secondary Cell Wall Biosynthesis in Maize
Journal of Plant Biology (2024)
-
Fine mapping of a new common bean anthracnose resistance gene (Co-18) to the proximal end of Pv10 in Indian landrace KRC-5
Theoretical and Applied Genetics (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.