Article
|
Open Access
Featured
-
-
Article
| Open Access Multi-species host range of staphylococcal phages isolated from wastewater
Multi-species host range of staphylococcal phages isolated from wastewater
The host range of bacteriophages defines their impact on bacterial ecology and diversity. Here, Göller et al. isolate 94 staphylococcal phages from wastewater and determine their host range on 117 staphylococci from 29 species, revealing a predominant multi-species host range and thus great potential for horizontal gene transfer.
- Pauline C. Göller
- , Tabea Elsener
- & Elena Gómez-Sanz
-
Article
| Open Access The origin and impeded dissemination of the DNA phosphorothioation system in prokaryotes
The origin and impeded dissemination of the DNA phosphorothioation system in prokaryotes
Phosphorothioate (PT) modification by the dnd gene cluster is the first identified DNA backbone modification and has been shown to constitute a multifunctional epigenetic system. Despite a variety of advantages for hosting dnd systems, these systems are surprisingly distributed sporadically among contemporary microbial genomes. To address this ecological paradox, Jian et al. systematically investigated the occurrence and phylogeny of dnd systems in prokaryotes, and provided evidence to suggest that dnd systems have originated in ancient Cyanobacteria (probably Nostocales) after the Great Oxygenation Event.
- Huahua Jian
- , Guanpeng Xu
- & Xiang Xiao
-
Article
| Open Access Community and single cell analyses reveal complex predatory interactions between bacteria in high diversity systems
Community and single cell analyses reveal complex predatory interactions between bacteria in high diversity systems
Studying the role of predator–prey interactions in food-web stability and species coexistence in the environment is arduous. Here, Cohen et al. use a combination of community and single-cell analyses to show that bacterial predators can regulate prey populations in the species-rich environments of wastewater treatment plants.
- Yossi Cohen
- , Zohar Pasternak
- & Edouard Jurkevitch
-
Article
| Open Access Prokaryotic viruses impact functional microorganisms in nutrient removal and carbon cycle in wastewater treatment plants
Prokaryotic viruses impact functional microorganisms in nutrient removal and carbon cycle in wastewater treatment plants
Activated sludge (AS) systems in wastewater treatment plants (WWTPs) contain high concentration of viruses. Here, the authors apply a systematic metagenomic pipeline and retrieve a catalogue of around 50,000 prokaryotic viruses from samples of six WWTPs, revealing a large and uncharacterized viral diversity in AS communities.
- Yiqiang Chen
- , Yulin Wang
- & Tong Zhang
-
Article
| Open Access Decline in plankton diversity and carbon flux with reduced sea ice extent along the Western Antarctic Peninsula
Decline in plankton diversity and carbon flux with reduced sea ice extent along the Western Antarctic Peninsula
Over the past century, the Western Antarctic Peninsula has experienced rapid warming and a substantial loss of sea ice with important implications for plankton biodiversity and carbon cycling. Using a 5-year DNA metabarcoding dataset, this study assesses how interannual variability in sea-ice conditions impacts biodiversity and biological carbon fluxes in this region.
- Yajuan Lin
- , Carly Moreno
- & Nicolas Cassar
-
Article
| Open Access Purple sulfur bacteria fix N2 via molybdenum-nitrogenase in a low molybdenum Proterozoic ocean analogue
Purple sulfur bacteria fix N2 via molybdenum-nitrogenase in a low molybdenum Proterozoic ocean analogue
N2 fixation was key to the expansion of life on Earth, but which organisms fixed N2 and if Mo-nitrogenase was functional in the low Mo early ocean is unknown. Here, the authors show that purple sulfur bacteria fix N2 using Mo-nitrogenase in a Proterozoic ocean analogue, despite low Mo conditions.
- Miriam Philippi
- , Katharina Kitzinger
- & Marcel M. M. Kuypers
-
Article
| Open Access Global distribution patterns of marine nitrogen-fixers by imaging and molecular methods
Global distribution patterns of marine nitrogen-fixers by imaging and molecular methods
Nitrogen fixation by diazotrophs is critical for marine primary production. Using Tara Oceans datasets, this study combines a quantitative image analysis pipeline with metagenomic mining to provide an improved global overview of diazotroph abundance, diversity and distribution.
- Juan José Pierella Karlusich
- , Eric Pelletier
- & Rachel A. Foster
-
Article
| Open Access A distinct growth physiology enhances bacterial growth under rapid nutrient fluctuations
A distinct growth physiology enhances bacterial growth under rapid nutrient fluctuations
Here the authors use microfluidics and single-cell microscopy to quantify the growth dynamics of individual E. coli cells exposed to nutrient fluctuations with periods as short as 30 seconds, finding that nutrient fluctuations reduce growth rates up to 50% compared to a steady nutrient delivery of equal average concentration, implying that temporal variability is an important parameter in bacterial growth.
- Jen Nguyen
- , Vicente Fernandez
- & Roman Stocker
-
Article
| Open Access Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics
Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics
Advances in omics provide a tool to understand mechanisms for plant–microbial interactions under stress. Here the authors apply genome-resolved metagenomics to investigate sorghum and its microbiome responses to drought, identifying an unexpected role of iron metabolism.
- Ling Xu
- , Zhaobin Dong
- & Devin Coleman-Derr
-
Article
| Open Access Ecology and molecular targets of hypermutation in the global microbiome
Ecology and molecular targets of hypermutation in the global microbiome
Here, the authors report a large-scale comparative analysis of <30,000 Diversity-Generating Retroelements (DGRs) across ~9000 metagenomes (representing diverse taxa and biomes), to identify patterns in terms of prevalence and activity. Combined with examination of longitudinal data on <100 metagenomes part of time series, they demonstrate that DGRs are broadly and consistently active, implying an important role in microbiota ecology and evolution.
- Simon Roux
- , Blair G. Paul
- & Emiley A. Eloe-Fadrosh
-
Article
| Open Access Decrypting bacterial polyphenol metabolism in an anoxic wetland soil
Decrypting bacterial polyphenol metabolism in an anoxic wetland soil
It is thought that polyphenols inhibit organic matter decomposition in soils devoid of oxygen. Here the authors use metabolomics and genome-resolved metaproteomics to provide experimental evidence of polyphenol biodegradation and maintained soil microbial community metabolism despite anoxia.
- Bridget B. McGivern
- , Malak M. Tfaily
- & Kelly C. Wrighton
-
Article
| Open Access Metabolic capabilities mute positive response to direct and indirect impacts of warming throughout the soil profile
Metabolic capabilities mute positive response to direct and indirect impacts of warming throughout the soil profile
There is much uncertainty on the response of soil microbial communities to warming, particularly in the subsoil. Here, the authors investigate microbial community and metabolism response to 4.5 years of whole-profile soil warming, finding depth-dependent effects and elevated subsoil microbial respiration.
- Nicholas C. Dove
- , Margaret S. Torn
- & Neslihan Taş
-
Article
| Open Access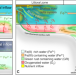 Diurnal Fe(II)/Fe(III) cycling and enhanced O2 production in a simulated Archean marine oxygen oasis
Diurnal Fe(II)/Fe(III) cycling and enhanced O2 production in a simulated Archean marine oxygen oasis
Cyanobacterial photosynthesis is thought to have oxygenated Earth’s atmosphere during the Great Oxidation Event, but these organisms had to overcome the toxic effects of iron. Here the authors simulate Archaean conditions in Cyanobacterial cultures and find that gas exchange and rust formation alleviated iron toxicity.
- A. J. Herrmann
- , J. Sorwat
- & M. M. Gehringer
-
Article
| Open Access Connecting structure to function with the recovery of over 1000 high-quality metagenome-assembled genomes from activated sludge using long-read sequencing
Connecting structure to function with the recovery of over 1000 high-quality metagenome-assembled genomes from activated sludge using long-read sequencing
Microbes play key roles in wastewater treatment. Here, Singleton et al. use long-read and short-read sequencing to recover 1083 high-quality metagenome-assembled genomes from 23 wastewater treatment plants, and combine this information with amplicon data, Raman microspectroscopy and FISH to reveal functionally important lineages.
- Caitlin M. Singleton
- , Francesca Petriglieri
- & Mads Albertsen
-
Article
| Open Access C-STABILITY an innovative modeling framework to leverage the continuous representation of organic matter
C-STABILITY an innovative modeling framework to leverage the continuous representation of organic matter
Soil organic matter (SOM) is a huge sink of carbon, but the varied flux dynamics are challenging to predict. Here, the authors present a new model with the complexities of SOM cycling, including parameters for substrate accessibility, microbe diversity, and enzymatic substrate depolymerization.
- Julien Sainte-Marie
- , Matthieu Barrandon
- & Delphine Derrien
-
Comment
| Open Access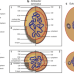 Cultivation of elusive microbes unearthed exciting biology
Cultivation of elusive microbes unearthed exciting biology
Many newly-discovered microbial phyla have been studied solely by cultivation-independent techniques such as metagenomics. Much of their biology thus remains elusive, because the organisms have not yet been isolated and grown in the lab. Katayama et al. lift the curtain on some intriguing biology by cultivating and studying bacteria from the elusive OP9 phylum (Atribacterota).
- Muriel C. F. van Teeseling
- & Christian Jogler
-
Article
| Open Access Integration of time-series meta-omics data reveals how microbial ecosystems respond to disturbance
Integration of time-series meta-omics data reveals how microbial ecosystems respond to disturbance
Herold et al. present an integrated meta-omics framework to investigate how mixed microbial communities, such as oleaginous bacterial populations in biological wastewater treatment plants, respond with distinct adaptation strategies to disturbances. They show that community resistance and resilience are a function of phenotypic plasticity and niche complementarity.
- Malte Herold
- , Susana Martínez Arbas
- & Paul Wilmes
-
Article
| Open Access A bacterial endophyte exploits chemotropism of a fungal pathogen for plant colonization
A bacterial endophyte exploits chemotropism of a fungal pathogen for plant colonization
Soil-borne fungal pathogens use chemotropism and extracellular pH alkalinisation to reach and penetrate plant roots. Here, Palmieri et al. show that soil endophytic bacteria swim along fungal hyphae to colonize plant roots and protect host plants by modulating the pH of the rhizosphere.
- Davide Palmieri
- , Stefania Vitale
- & David Turrà
-
Article
| Open Access Bacteria are important dimethylsulfoniopropionate producers in marine aphotic and high-pressure environments
Bacteria are important dimethylsulfoniopropionate producers in marine aphotic and high-pressure environments
Dimethylsulfoniopropionate (DMSP) is an osmolyte produced by marine microbes that plays an important role in nutrient cycling and atmospheric chemistry. Here the authors go to the Mariana Trench—the deepest point in the ocean—and find bacteria are key DMSP producers, and that DMSP has a role in protection against high pressure.
- Yanfen Zheng
- , Jinyan Wang
- & Xiao-Hua Zhang
-
Article
| Open Access Synthetic microbial communities of heterotrophs and phototrophs facilitate sustainable growth
Synthetic microbial communities of heterotrophs and phototrophs facilitate sustainable growth
Successful application of microbial community for bioproduction relies on the selection of appropriate heterotroph and phototroph partners. Here, the authors construct community metabolic models to guide strain selection and experimentally validate metabolic exchanges that sustain the heterotrophs in minimal media.
- Cristal Zuñiga
- , Tingting Li
- & Karsten Zengler
-
Article
| Open Access Aerobic microbial life persists in oxic marine sediment as old as 101.5 million years
Aerobic microbial life persists in oxic marine sediment as old as 101.5 million years
The discovery of aerobic microbial communities in nutrient-poor sediments below the seafloor begs the question of the mechanisms for their persistence. Here the authors investigate subseafloor sediment in the South Pacific Gyre abyssal plain, showing that aerobic microbial life can be revived and retain metabolic potential even from 101.5 Ma-old sediment.
- Yuki Morono
- , Motoo Ito
- & Fumio Inagaki
-
Article
| Open Access Microbial diversity drives carbon use efficiency in a model soil
Microbial diversity drives carbon use efficiency in a model soil
Microbial carbon use efficiency has an important role in soil C cycling. Here the authors test the interactive effects of temperature and moisture and manipulate microbial community composition in soil microcosms, showing a positive relationship between microbial diversity and CUE that is contingent on abiotic conditions.
- Luiz A. Domeignoz-Horta
- , Grace Pold
- & Kristen M. DeAngelis
-
Article
| Open Access Bacterial symbionts support larval sap feeding and adult folivory in (semi-)aquatic reed beetles
Bacterial symbionts support larval sap feeding and adult folivory in (semi-)aquatic reed beetles
Symbiotic microbes in insects can enable their hosts to access untapped nutritional resources. Here, the authors show that symbiotic bacteria in reed beetles can provide essential amino acids to sap-feeding larvae and help leaf-feeding adults to degrade pectin, respectively.
- Frank Reis
- , Roy Kirsch
- & Martin Kaltenpoth
-
Article
| Open Access Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients
Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients
Here, the authors sample air and surfaces in hospital rooms of COVID-19 patients, detect SARS-CoV-2 RNA in air samples of two of three tested airborne infection isolation rooms, and find surface contamination in 66.7% of tested rooms during the first week of illness and 20% beyond the first week of illness.
- Po Ying Chia
- , Kristen Kelli Coleman
- & Daniela Moses
-
Article
| Open Access Genome-scale metabolic reconstruction of the symbiosis between a leguminous plant and a nitrogen-fixing bacterium
Genome-scale metabolic reconstruction of the symbiosis between a leguminous plant and a nitrogen-fixing bacterium
The association between leguminous plants and rhizobial bacteria is a paradigmatic example of a symbiosis driven by metabolic exchanges. Here, diCenzo et al. report the reconstruction and modelling of a genome-scale metabolic network of the plant Medicago truncatula nodulated by the bacterium Sinorhizobium meliloti.
- George C. diCenzo
- , Michelangelo Tesi
- & Marco Fondi
-
Article
| Open Access Extracellular electron transfer-dependent anaerobic oxidation of ammonium by anammox bacteria
Extracellular electron transfer-dependent anaerobic oxidation of ammonium by anammox bacteria
Bacteria capable of anaerobic ammonium oxidation (anammox) produce half of the nitrogen gas in the atmosphere, but much of their physiology is still unknown. Here the authors show that anammox bacteria are capable of a novel mechanism of ammonium oxidation using extracellular electron transfer.
- Dario R. Shaw
- , Muhammad Ali
- & Pascal E. Saikaly
-
Article
| Open Access Cable bacteria reduce methane emissions from rice-vegetated soils
Cable bacteria reduce methane emissions from rice-vegetated soils
Rice paddies are a major source of the Earth’s atmospheric methane, making these important food crops potent contributors to greenhouse gas emissions. Here the authors show that inoculation of paddies with a particular bacterium could significantly curb methane production.
- Vincent V. Scholz
- , Rainer U. Meckenstock
- & Nils Risgaard-Petersen
-
Article
| Open Access Single cell analyses reveal contrasting life strategies of the two main nitrifiers in the ocean
Single cell analyses reveal contrasting life strategies of the two main nitrifiers in the ocean
Ammonia oxidizing archaea and Nitrospinae are the main known nitrifiers in the ocean, but the much greater abundance of the former is puzzling. Here, the authors show that differences in mortality, rather than thermodynamics, cell size or biomass yield, explain the discrepancy, without the need to invoke yet undiscovered, abundant nitrite oxidizers.
- Katharina Kitzinger
- , Hannah K. Marchant
- & Marcel M. M. Kuypers
-
Article
| Open Access The oomycete Lagenisma coscinodisci hijacks host alkaloid synthesis during infection of a marine diatom
The oomycete Lagenisma coscinodisci hijacks host alkaloid synthesis during infection of a marine diatom
Flagellated oomycetes frequently infect unicellular algae, thus limiting their proliferation. Here, the authors show that an oomycete rewires the metabolome of a marine bloom-forming diatom, thereby promoting infection success.
- Marine Vallet
- , Tim U. H. Baumeister
- & Georg Pohnert
-
Article
| Open Access A shared core microbiome in soda lakes separated by large distances
A shared core microbiome in soda lakes separated by large distances
Alkaline lakes have some of the highest productivity rates in freshwater ecosystems. Here the authors report amplicon, metagenome, and proteome sequencing from microbial mat communities of four alkaline lakes in Canada, and compare these lakes to central Asian soda lakes, revealing a shared core microbiome despite the geographical distance.
- Jackie K. Zorz
- , Christine Sharp
- & Marc Strous
-
Article
| Open Access Changes in the metabolic potential of the sponge microbiome under ocean acidification
Changes in the metabolic potential of the sponge microbiome under ocean acidification
Anthropogenic CO2 emissions are causing ocean acidification, which can affect the physiology of some organisms. Here, Botté et al. use metagenomics to show differences in metabolic potential between sponge microbiomes sampled at a shallow volcanic CO2 seep and those from nearby control sites.
- Emmanuelle S. Botté
- , Shaun Nielsen
- & Nicole S. Webster
-
Article
| Open Access Assembly and seasonality of core phyllosphere microbiota on perennial biofuel crops
Assembly and seasonality of core phyllosphere microbiota on perennial biofuel crops
Microbial communities of plant leaf surfaces are ecologically important, but how they assemble and vary in time is unclear. Here, the authors identify core leaf microbiomes and seasonal patterns for two biofuel crops and show with source-sink models that soil is a reservoir of phyllosphere diversity.
- Keara L. Grady
- , Jackson W. Sorensen
- & Ashley Shade
-
Article
| Open Access Space Station conditions are selective but do not alter microbial characteristics relevant to human health
Space Station conditions are selective but do not alter microbial characteristics relevant to human health
The International Space Station is a unique habitat for humans and microbes. Here, Mora et al. analyze microbial communities from several areas aboard, finding similarities with those of ground-based indoor environments, as well as adaptations towards biofilm formation but not necessarily relevant to human health.
- Maximilian Mora
- , Lisa Wink
- & Christine Moissl-Eichinger
-
Article
| Open Access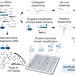 Uncovering the biosynthetic potential of rare metagenomic DNA using co-occurrence network analysis of targeted sequences
Uncovering the biosynthetic potential of rare metagenomic DNA using co-occurrence network analysis of targeted sequences
Soil microorganisms are a rich source of bioactive molecules. Here, the authors present a targeted sequencing workflow that reconstructs the clustered organization of biosynthetic domains in metagenomic libraries from amplicon data, thus guiding the discovery of novel metabolites from rare members of the soil microbiome.
- Vincent Libis
- , Niv Antonovsky
- & Sean F. Brady
-
Article
| Open Access Diversity-triggered deterministic bacterial assembly constrains community functions
Diversity-triggered deterministic bacterial assembly constrains community functions
The role of microbial β-diversity in soil ecosystem function is not well-studied. Here, the authors use genetic data to show that microbial α-diversity levels may have impacts on stochastic/deterministic assembly processes and functions of soil microbiome.
- Weibing Xun
- , Wei Li
- & Ruifu Zhang
-
Article
| Open Access Probing the active fraction of soil microbiomes using BONCAT-FACS
Probing the active fraction of soil microbiomes using BONCAT-FACS
Standard DNA-based analyses of microbial communities cannot distinguish between active microbes and dead or dormant cells. Here, Couradeau et al. use BONCAT (bioorthogonal non-canonical amino acid tagging), flow cytometry, and 16S rRNA gene amplicon sequencing to identify active microbial cells in soils.
- Estelle Couradeau
- , Joelle Sasse
- & Trent R. Northen
-
Article
| Open Access Phylogenetic conservation of bacterial responses to soil nitrogen addition across continents
Phylogenetic conservation of bacterial responses to soil nitrogen addition across continents
Developing a predictive understanding of bacterial community responses to environmental change is an ongoing challenge. Here, Isobe et al. reanalyze data on soil microbial responses to nitrogen addition across 5 continents, finding that responses are predictable based on phylogeny.
- Kazuo Isobe
- , Steven D. Allison
- & Jennifer B. H. Martiny
-
Article
| Open Access A few Ascomycota taxa dominate soil fungal communities worldwide
A few Ascomycota taxa dominate soil fungal communities worldwide
Soil fungi play essential roles in ecosystems worldwide. Here, the authors sequence and analyze 235 soil samples collected from across the globe, and identify dominant fungal taxa and their associated environmental attributes.
- Eleonora Egidi
- , Manuel Delgado-Baquerizo
- & Brajesh K. Singh
-
Article
| Open Access Low yield and abiotic origin of N2O formed by the complete nitrifier Nitrospira inopinata
Low yield and abiotic origin of N2O formed by the complete nitrifier Nitrospira inopinata
Ammonia-oxidizing bacteria and archaea are major producers of the gases nitrous oxide and nitric oxide. Here, Kits et al. show that a complete ammonia-oxidizing (comammox) bacterium emits nitrous oxide at levels that are comparable to those produced by ammonia-oxidizing archaea.
- K. Dimitri Kits
- , Man-Young Jung
- & Holger Daims
-
Article
| Open Access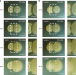 The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization
The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization
Pseudomonas and Bacillus can promote plant growth but their mutual interactions are unclear. Here, the authors show that the extracellular matrix protects Bacillus colonies from infiltration by Pseudomonas cells, while the Pseudomonas type VI secretion system stimulates Bacillus sporulation.
- Carlos Molina-Santiago
- , John R. Pearson
- & Diego Romero
-
Article
| Open Access Microbial and metabolic succession on common building materials under high humidity conditions
Microbial and metabolic succession on common building materials under high humidity conditions
Microbes inhabit built environments and could contribute to degradation of surfaces especially in damp conditions. Here the authors explore how communities of microbes and their metabolites affect four types of built surfaces under varying environmental conditions.
- Simon Lax
- , Cesar Cardona
- & Jack A. Gilbert
-
Article
| Open Access A new type of DNA phosphorothioation-based antiviral system in archaea
A new type of DNA phosphorothioation-based antiviral system in archaea
The prokaryote defence system Dnd relies on phosphorothioation (PT) of DNA backbone to distinguish between self and non-self DNA. Here, Xiong et al. describe a previously uncharacterized PT-based antiviral system that is independent of the canonical Dnd and is widespread in Archaea and Bacteria.
- Lei Xiong
- , Siyi Liu
- & Shi Chen
-
Article
| Open Access Colonization of the human gut by bovine bacteria present in Parmesan cheese
Colonization of the human gut by bovine bacteria present in Parmesan cheese
Some microorganisms may be transferred across the food production chain and, potentially, colonize the human gut. Here, Milani et al. provide strain-level evidence supporting that dairy cattle-associated bacteria can be transferred to the human gut via consumption of Parmesan cheese.
- Christian Milani
- , Sabrina Duranti
- & Francesca Turroni
-
Article
| Open Access Nitric oxide-dependent anaerobic ammonium oxidation
Nitric oxide-dependent anaerobic ammonium oxidation
Anammox bacteria couple nitrite reduction to ammonium oxidation, with nitric oxide (NO) and hydrazine as intermediates, and produce N2 and nitrate. Here, Hu et al. show that an anammox bacterium can grow in the absence of nitrite by coupling ammonium oxidation to NO reduction, producing only N2.
- Ziye Hu
- , Hans J. C. T. Wessels
- & Boran Kartal
-
Article
| Open Access Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage
Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage
Obtaining data on antimicrobial resistance (AMR) from healthy human populations is difficult. Here, Hendriksen et al. use metagenomic analysis to obtain AMR data from untreated sewage from 79 sites in 60 countries, finding correlations with socio-economic, health and environmental factors.
- Rene S. Hendriksen
- , Patrick Munk
- & Frank M. Aarestrup
-
Article
| Open Access Man-made microbial resistances in built environments
Man-made microbial resistances in built environments
The environmental microbiota can have important implications for our well-being. Here, the authors describe the composition of microbiomes from diverse buildings, including samples from clinical environments, and show that cleaner environments are associated with a loss of microbial diversity and an increase in genes associated with antibiotic resistance.
- Alexander Mahnert
- , Christine Moissl-Eichinger
- & Gabriele Berg
-
Article
| Open Access Marine biofilms constitute a bank of hidden microbial diversity and functional potential
Marine biofilms constitute a bank of hidden microbial diversity and functional potential
Previous surveys of global ocean microbial diversity have focused on planktonic microbes. Here, Zhang et al. use metagenomics to study biofilm-forming marine microbes, increasing the known microbial diversity in the oceans by more than 20% and revealing new biosynthetic gene clusters and CRISPR-Cas systems.
- Weipeng Zhang
- , Wei Ding
- & Pei-Yuan Qian
-
Article
| Open Access Metabolic diversity within the globally abundant Marine Group II Euryarchaea offers insight into ecological patterns
Metabolic diversity within the globally abundant Marine Group II Euryarchaea offers insight into ecological patterns
The physiology and ecology of many yet-uncultured microorganisms, such as the Marine Group II Euryarchaea (MGII), is unclear. Here, Benjamin Tully analyses 250 MGII genomes, identifies 17 distinct subclades, and provides a detailed view of the metabolic potential and distribution of these archaea.
- Benjamin J. Tully
-
Article
| Open Access Metaepigenomic analysis reveals the unexplored diversity of DNA methylation in an environmental prokaryotic community
Metaepigenomic analysis reveals the unexplored diversity of DNA methylation in an environmental prokaryotic community
Our knowledge of DNA methylation systems in prokaryotes is mostly limited to those of culturable microbes. Here, Hiraoka et al. analyse DNA methylation patterns in metagenomic data from a microbial community, revealing new methylated motifs and experimentally validating the methyltransferases’ specificities.
- Satoshi Hiraoka
- , Yusuke Okazaki
- & Wataru Iwasaki

 Phylogenetically and functionally diverse microorganisms reside under the Ross Ice Shelf
Phylogenetically and functionally diverse microorganisms reside under the Ross Ice Shelf