Featured
-
-
Article
| Open Access Cooperative assembly of p97 complexes involved in replication termination
Cooperative assembly of p97 complexes involved in replication termination
This study describes how p97Ufd1-Npl4 and the UBA-UBX protein Ubxn7 disassemble vertebrate replisomes during replication termination, and it provides novel insights into how p97 complexes assemble with UBA-UBX proteins on ubiquitylated substrates
- Olga V. Kochenova
- , Sirisha Mukkavalli
- & Johannes C. Walter
-
Article
| Open Access Profilin-1 regulates DNA replication forks in a context-dependent fashion by interacting with SNF2H and BOD1L
Profilin-1 regulates DNA replication forks in a context-dependent fashion by interacting with SNF2H and BOD1L
Subcellular localization plays an important yet underappreciated role in protein functions. Here the authors defined novel and context-dependent nuclear functions of the actin-binding factor profilin-1 in DNA replication fork dynamics and stability.
- Cuige Zhu
- , Mari Iwase
- & Jieya Shao
-
Article
| Open Access Engineered helicase replaces thermocycler in DNA amplification while retaining desired PCR characteristics
Engineered helicase replaces thermocycler in DNA amplification while retaining desired PCR characteristics
PCR is an essential method for the amplification and manipulation of nucleic acids, but the requirement for a thermocycler limits access. Here, authors engineer a helicase to replace the heating step of PCR with enzymatic unwinding, allowing the isothermal amplification of fragments up to 6 kb.
- Momčilo Gavrilov
- , Joshua Y. C. Yang
- & Taekjip Ha
-
Article
| Open Access Solving the MCM paradox by visualizing the scaffold of CMG helicase at active replisomes
Solving the MCM paradox by visualizing the scaffold of CMG helicase at active replisomes
For several decades the MCM2-7 proteins, the core of the DNA replicative helicase, eluded detection at DNA replication sites. Here, the authors solve this conundrum by gene editing, which enables visualization of replication dynamics in living cells.
- Hana Polasek-Sedlackova
- , Thomas C. R. Miller
- & Jiri Lukas
-
Article
| Open Access ISG15 conjugation to proteins on nascent DNA mitigates DNA replication stress
ISG15 conjugation to proteins on nascent DNA mitigates DNA replication stress
DNA replication stress can result in genome instability. Here the authors show that the ubiquitin like modifier protein, ISG15, important during the innate immune response, acts at replication forks to mitigate DNA replication stress.
- Christopher P. Wardlaw
- & John H. J. Petrini
-
Article
| Open Access Lagging strand gap suppression connects BRCA-mediated fork protection to nucleosome assembly through PCNA-dependent CAF-1 recycling
Lagging strand gap suppression connects BRCA-mediated fork protection to nucleosome assembly through PCNA-dependent CAF-1 recycling
Efficient DNA replication is crucial for genome stability. Here, Thakar et al. report that accumulation of lagging strand ssDNA gaps during replication interferes with nucleosome assembly and drives replication fork degradation in BRCA-deficient cells.
- Tanay Thakar
- , Ashna Dhoonmoon
- & George-Lucian Moldovan
-
Article
| Open Access MAD2L2 promotes replication fork protection and recovery in a shieldin-independent and REV3L-dependent manner
MAD2L2 promotes replication fork protection and recovery in a shieldin-independent and REV3L-dependent manner
MAD2L2 – as a member of the shieldin complex - counteracts resection during DNA repair. Here the authors demonstrate that MAD2L2 protects stalled replication forks from excessive resection, in a shieldin-independent and REV3L-dependent manner.
- Inés Paniagua
- , Zainab Tayeh
- & Jacqueline J. L. Jacobs
-
Article
| Open Access Nucleosome-directed replication origin licensing independent of a consensus DNA sequence
Nucleosome-directed replication origin licensing independent of a consensus DNA sequence
Most eukaryotes do not use a consensus DNA sequence as binding sites for the origin recognition complex (ORC) to initiate DNA replication, however budding yeast do. Here the authors show S. cerevisiae ORC can bind nucleosomes near nucleosome-free regions and recruit replicative helicases to form a pre-replication complex independent of the DNA sequence.
- Sai Li
- , Michael R. Wasserman
- & Shixin Liu
-
Article
| Open Access The mechanism of replication stalling and recovery within repetitive DNA
The mechanism of replication stalling and recovery within repetitive DNA
DNA replication of repetitive sequences was recreated in a test tube using purified components. DNA alone was sufficient to induce stalling. Both stalling and recovery were dictated by the capacity of DNA to fold into unusual secondary structures.
- Corella S. Casas-Delucchi
- , Manuel Daza-Martin
- & Gideon Coster
-
Article
| Open Access A plasmid system with tunable copy number
A plasmid system with tunable copy number
The range of available copy numbers for cloning vectors is largely restricted to the handful of ORIs that have been isolated from plasmids found in nature. Here the authors introduce a plasmid system that allow for the continuous, finely-tuned control of plasmid copy number between 1 and 800 copies per cell.
- Miles V. Rouches
- , Yasu Xu
- & Guillaume Lambert
-
Article
| Open Access Dynamic interaction of BRCA2 with telomeric G-quadruplexes underlies telomere replication homeostasis
Dynamic interaction of BRCA2 with telomeric G-quadruplexes underlies telomere replication homeostasis
G-quadruplex (G4) can be formed in telomeric DNA. Here the authors show that BRCA2 interacts with telomere G4 structure generated during telomere replication, protecting telomere from nuclease attack.
- Junyeop Lee
- , Keewon Sung
- & Hyunsook Lee
-
Article
| Open Access Genome-wide mapping of individual replication fork velocities using nanopore sequencing
Genome-wide mapping of individual replication fork velocities using nanopore sequencing
Theulot et al. introduce NanoForkSpeed, a nanopore sequencing-based method to map individual replication fork velocities on entire genomes. NFS shows that fork speed is uniform across yeast chromosomes except for a marked slowdown at pausing sites.
- Bertrand Theulot
- , Laurent Lacroix
- & Benoît Le Tallec
-
Article
| Open Access Stable inheritance of H3.3-containing nucleosomes during mitotic cell divisions
Stable inheritance of H3.3-containing nucleosomes during mitotic cell divisions
How nucleosome assembly of parental histones is regulated following DNA replication is still an open question. Here the authors show that unlike deposition of new histones H3.1 and H3.3 that utilizes different histone chaperones, parental H3.1 and H3.3 are both stably inherited during mitotic cell division in mouse embryonic stem cells, and this involves histone chaperones Mcm2, Pole3 and Pole4.
- Xiaowei Xu
- , Shoufu Duan
- & Zhiguo Zhang
-
Article
| Open Access Rad51-mediated replication of damaged templates relies on monoSUMOylated DDK kinase
Rad51-mediated replication of damaged templates relies on monoSUMOylated DDK kinase
Joseph et al. reveal that monoSUMOylated DDK kinase, implicated in replication initiation, acts with Rad51 recombinase to prevent replication fork uncoupling and to mediate recombination-dependent gap-filling in the presence of genotoxic stress.
- Chinnu Rose Joseph
- , Sabrina Dusi
- & Dana Branzei
-
Article
| Open Access In crystallo observation of three metal ion promoted DNA polymerase misincorporation
In crystallo observation of three metal ion promoted DNA polymerase misincorporation
By observing DNA polymerase misincorporation with time-resolved crystallography, the authors visualize three-metal ion dependent polymerase catalysis and identify A-site metal-mediated primer alignment as a key step in nucleotide discrimination.
- Caleb Chang
- , Christie Lee Luo
- & Yang Gao
-
Article
| Open Access Kronos scRT: a uniform framework for single-cell replication timing analysis
Kronos scRT: a uniform framework for single-cell replication timing analysis
A scalable approach to explore DNA replication in single cells reveals that although aneuploidy does not have a major impact on the pattern of replication, different cell types and sub-populations display distinguished replication paths.
- Stefano Gnan
- , Joseph M. Josephides
- & Chun-Long Chen
-
Article
| Open Access USP1-trapping lesions as a source of DNA replication stress and genomic instability
USP1-trapping lesions as a source of DNA replication stress and genomic instability
Here the authors provide mechanistic insights into how auto-cleavage of the USP1 deubiquitinase regulates DNA replication and genome stability. Implications for the targeting of USP1 activity via protein-DNA trapping in cancer therapy are discussed.
- Kate E. Coleman
- , Yandong Yin
- & Tony T. Huang
-
Article
| Open Access Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase
Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase
The Dbf4-dependent kinase Cdc7 (DDK) is essential for eukaryotic DNA replication. Here, the authors present a series of cryo-EM structures elucidating the versatility of this kinase in exerting an ordered phosphorylation of its essential target to promote replication initiation.
- Jiaxuan Cheng
- , Ningning Li
- & Yuanliang Zhai
-
Article
| Open Access Cryo-EM structure of translesion DNA synthesis polymerase ζ with a base pair mismatch
Cryo-EM structure of translesion DNA synthesis polymerase ζ with a base pair mismatch
The structure of mismatched DNA-Polζ ternary complex provides a basis for understanding what makes Polζ adept at extending DNA synthesis past mismatched base pairs.
- Radhika Malik
- , Robert E. Johnson
- & Aneel K. Aggarwal
-
Article
| Open Access A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6
A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6
Cryo-EM structures of S. cerevisiae ORC bound to DNA and Cdc6 reveal an autoinhibited conformation and suggest a mechanism of origin licensing control in response to CDK phosphorylation.
- Jan Marten Schmidt
- , Ran Yang
- & Franziska Bleichert
-
Article
| Open Access The combined DNA and RNA synthetic capabilities of archaeal DNA primase facilitate primer hand-off to the replicative DNA polymerase
The combined DNA and RNA synthetic capabilities of archaeal DNA primase facilitate primer hand-off to the replicative DNA polymerase
DNA primases initiate a short primer before handing off to DNA polymerases to continue replication. Here the authors reveal a unique ability of archaeal primases to first synthesize RNA before stochastically incorporating a deoxyribonucleotide and further extending the primer as DNA.
- Mark D. Greci
- , Joseph D. Dooher
- & Stephen D. Bell
-
Article
| Open Access BRCA1 deficiency specific base substitution mutagenesis is dependent on translesion synthesis and regulated by 53BP1
BRCA1 deficiency specific base substitution mutagenesis is dependent on translesion synthesis and regulated by 53BP1
Loss of BRCA1 or BRCA2 results in genomic instability; however most studies have focused on the role of these proteins in double-strand break repair. Here the authors coupled cell line genetics and whole genome sequencing to investigate the formation of base substitutions and short indels in BRCA1-deficient cells, revealing a role for translesion DNA synthesis regulated by 53BP1.
- Dan Chen
- , Judit Z. Gervai
- & Dávid Szüts
-
Article
| Open Access The genetic architecture of DNA replication timing in human pluripotent stem cells
The genetic architecture of DNA replication timing in human pluripotent stem cells
The genetic basis of how cells replicate their DNA is not well understood. Here, the authors identify >1000 genetic elements that control human replication and reveal a complex epigenetic system that regulates replication origin activities.
- Qiliang Ding
- , Matthew M. Edwards
- & Amnon Koren
-
Article
| Open Access WRN helicase safeguards deprotected replication forks in BRCA2-mutated cancer cells
WRN helicase safeguards deprotected replication forks in BRCA2-mutated cancer cells
The tumor suppressor BRCA2 protects stalled DNA replication forks from unrestrained degradation; however the mechanism whereby unprotected stalled forks are preserved and restarted has remained elusive. Here the authors show that the WRN helicase promotes stalled fork recovery and limits fork hyper-degradation in the absence of BRCA2 protection.
- Arindam Datta
- , Kajal Biswas
- & Robert M. Brosh Jr
-
Article
| Open Access BRCA2 associates with MCM10 to suppress PRIMPOL-mediated repriming and single-stranded gap formation after DNA damage
BRCA2 associates with MCM10 to suppress PRIMPOL-mediated repriming and single-stranded gap formation after DNA damage
Tumor suppressor BRCA2 is known to stabilize and restart stalled DNA replication forks. Here the authors show that BRCA2 is recruited to the replication fork through its interaction with MCM10 and inhibits Primase-Polymerase-mediated repriming, lesion bypass and single strand DNA gap formation after DNA damage.
- Zhihua Kang
- , Pan Fu
- & Bing Xia
-
Article
| Open Access SPOP mutation induces replication over-firing by impairing Geminin ubiquitination and triggers replication catastrophe upon ATR inhibition
SPOP mutation induces replication over-firing by impairing Geminin ubiquitination and triggers replication catastrophe upon ATR inhibition
Geminin-Cdt1 plays essential roles in the regulation of DNA replication. Here the authors reveal that the CULLIN3 E3 ubiquitin ligase adaptor protein SPOP prevents DNA replication over-firing and genome instability by affecting Geminin ubiquitination.
- Jian Ma
- , Qing Shi
- & Haojie Huang
-
Article
| Open Access A novel lncRNA Discn fine-tunes replication protein A (RPA) availability to promote genomic stability
A novel lncRNA Discn fine-tunes replication protein A (RPA) availability to promote genomic stability
Various lncRNAs functions have been associated to replication stress and DNA double strand breaks in ESC. Here the authors reveal the lncRNA Discn-NCL-RPA axis pathway, which can regulate the availability of RPA pool for DNA replication stress response and repair.
- Lin Wang
- , Jingzheng Li
- & Ping Zheng
-
Article
| Open Access Unwinding of a DNA replication fork by a hexameric viral helicase
Unwinding of a DNA replication fork by a hexameric viral helicase
Replicative hexameric helicases are fundamental components of replisomes. Here the authors resolve a cryo-EM structure of the E1 helicase from papillomavirus bound to a DNA replication fork, providing insights into the mechanism of DNA unwinding by these hexameric enzymes.
- Abid Javed
- , Balazs Major
- & Elena V. Orlova
-
Article
| Open Access The RAD51 recombinase protects mitotic chromatin in human cells
The RAD51 recombinase protects mitotic chromatin in human cells
RAD51 is a well known player of DNA repair and homologous recombination. Here the authors reveal a function for RAD51 in protecting under-replicated DNA in mitotic human cells, promoting mitotic DNA synthesis (MiDAS) and successful chromosome segregation.
- Isabel E. Wassing
- , Emily Graham
- & Fumiko Esashi
-
Article
| Open Access RNF168-mediated localization of BARD1 recruits the BRCA1-PALB2 complex to DNA damage
RNF168-mediated localization of BARD1 recruits the BRCA1-PALB2 complex to DNA damage
The BRCA1-PALB2-BRCA2-RAD51 (BRCA1-P) complex is well known to play a fundamental role in DNA repair, but how the complex recruitment is regulated is still a matter of interest. Here the authors reveal mechanistic insights into RNF168 activity being responsible for PALB2 recruitment, through BARD1-BRCA1 during homologous recombination repair.
- John J. Krais
- , Yifan Wang
- & Neil Johnson
-
Article
| Open Access A transcription-based mechanism for oncogenic β-catenin-induced lethality in BRCA1/2-deficient cells
A transcription-based mechanism for oncogenic β-catenin-induced lethality in BRCA1/2-deficient cells
Germline mutations in BRCA1 or BRCA2 tumour suppressor genes predispose to different cancers, as does oncogene activation. Here the authors reveal that aberrant transcription of specific genes triggered by activation of the oncogene β-catenin causes replication failure and cell death in the context of BRCA1/2 deficiency.
- Rebecca A. Dagg
- , Gijs Zonderland
- & Madalena Tarsounas
-
Perspective
| Open Access Towards a synthetic cell cycle
Towards a synthetic cell cycle
A key feature of living cells is the cell cycle. In this Perspective, the authors explore attempts to recreate this process and what is still required for an integrated synthetic cell cycle.
- Lorenzo Olivi
- , Mareike Berger
- & John van der Oost
-
Article
| Open Access The human nucleoporin Tpr protects cells from RNA-mediated replication stress
The human nucleoporin Tpr protects cells from RNA-mediated replication stress
Tpr nucleoporin is known to be essential for nuclear transport and mitosis processes. Here the authors explore the link between Tpr and genome instability providing insights into the role of Tpr in safeguarding cells from RNA-mediated replication stress.
- Martin Kosar
- , Michele Giannattasio
- & Marco Foiani
-
Article
| Open Access RPA shields inherited DNA lesions for post-mitotic DNA synthesis
RPA shields inherited DNA lesions for post-mitotic DNA synthesis
Single-stranded DNA during DNA replication and repair in S/G2 needs protection by replication protein A (RPA). Here the authors reveal that RPA also shields inherited single-stranded DNA in G1, representing replication remnants from the previous cell cycle, to allow for post-mitotic DNA synthesis.
- Aleksandra Lezaja
- , Andreas Panagopoulos
- & Matthias Altmeyer
-
Article
| Open Access Mechanism of genome instability mediated by human DNA polymerase mu misincorporation
Mechanism of genome instability mediated by human DNA polymerase mu misincorporation
Pol μ performs gap-filling repair synthesis in the nonhomologous end joining (NHEJ) pathway. Here the authors provide crystal structures and kinetics of human Pol μ to reveal insights into the molecular mechanism of Pol μ during the process of dGTP misincorporation.
- Miao Guo
- , Yina Wang
- & Ye Zhao
-
Article
| Open Access BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage
BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage
BRCA1-mediated resolution of R-loops has previously been described. Here the authors reveal a functional association of BRCA1 with TERRA RNA at telomeres, which develops in an R-loop-, and a cell cycle-dependent manner.
- Jekaterina Vohhodina
- , Liana J. Goehring
- & David M. Livingston
-
Article
| Open Access Dynamics of replication origin over-activation
Dynamics of replication origin over-activation
DNA replication processes are often dysregulated in cancer. Here the authors analyse DNA synthesis patterns in cancer cells undergoing partial genome re-replication to reveal that re-replication exhibits aberrant replication fork dynamics and a skewed distribution of replication initiation that over-duplicates early-replicating genomic regions.
- Haiqing Fu
- , Christophe E. Redon
- & Mirit I. Aladjem
-
Article
| Open Access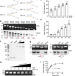 MIF is a 3’ flap nuclease that facilitates DNA replication and promotes tumor growth
MIF is a 3’ flap nuclease that facilitates DNA replication and promotes tumor growth
Replication stress is associated with cancer formation and progression. Here the authors reveal that the macrophage migration inhibitory factor (MIF) functions as 3’ flap nuclease involved in resolving replication stress affecting overall tumor progression.
- Yijie Wang
- , Yan Chen
- & Yingfei Wang
-
Article
| Open Access Mechanism of MRX inhibition by Rif2 at telomeres
Mechanism of MRX inhibition by Rif2 at telomeres
Different proteins localised at telomeres ensure chromosome end stability to prevent double strand-end break recognition. Here the authors provide new insight into how in S. cerevisiae the interaction between Rif2 and Rad50 inhibits MRX functions at telomeres.
- Florian Roisné-Hamelin
- , Sabrina Pobiega
- & Stéphane Marcand
-
Article
| Open Access Single-molecule imaging reveals replication fork coupled formation of G-quadruplex structures hinders local replication stress signaling
Single-molecule imaging reveals replication fork coupled formation of G-quadruplex structures hinders local replication stress signaling
In the genome, repetitive guanine-rich sequences have the potential to spontaneously fold into non-canonical DNA secondary structures known as G-quadruplex (G4). Using novel single-molecule imaging approaches, the authors reveal that G4 formation within active replication forks locally perturb replisome dynamics and damage response signaling, which require RPA and FANCJ for regulation.
- Wei Ting C. Lee
- , Yandong Yin
- & Eli Rothenberg
-
Article
| Open Access Smc5/6 functions with Sgs1-Top3-Rmi1 to complete chromosome replication at natural pause sites
Smc5/6 functions with Sgs1-Top3-Rmi1 to complete chromosome replication at natural pause sites
Smc5/6, part of the structural maintenance of chromosomes (SMC) family, plays roles in genome structural integrity. Here the authors reveal that Smc5/6 acts jointly with Top3 within the STR complex to mediate DNA replication completion at genomic natural pausing sites (NPSs).
- Sumedha Agashe
- , Chinnu Rose Joseph
- & Dana Branzei
-
Article
| Open Access DNA replication origins retain mobile licensing proteins
DNA replication origins retain mobile licensing proteins
Eukaryotic DNA replication is regulated to ensure copying of the genome (only) once per cell cycle. Here the authors, using optical trapping and confocal microscopy, demonstrate the dynamics of the origin recognition complex and subsequent intermediates that lead up to the loading of an MCM helicase onto DNA.
- Humberto Sánchez
- , Kaley McCluskey
- & Nynke H. Dekker
-
Article
| Open Access Stabilisation of half MCM ring by Cdt1 during DNA insertion
Stabilisation of half MCM ring by Cdt1 during DNA insertion
During pre-Replication Complex, eukaryotic cells load two MCMs into a head-to-head Double Hexamer around duplex DNA (DH). Here the authors preRC assembly assay with purified proteins to reveal insights into S. cerevisiae’s first steps that lead to the DH formation.
- Marina Guerrero-Puigdevall
- , Narcis Fernandez-Fuentes
- & Jordi Frigola
-
Article
| Open Access Chromosomal coordination and differential structure of asynchronous replicating regions
Chromosomal coordination and differential structure of asynchronous replicating regions
Most regions of the mammalian genome replicate both alleles in a synchronous manner, but some loci have been found to replicate asynchronously and the time of replication of each allele is different. Here the authors, by employing clonal mouse cells from a hybrid strain chart replication timing over the entire genome, using polymorphisms to distinguish between the paternal and maternal alleles.
- Britny Blumenfeld
- , Hagit Masika
- & Itamar Simon
-
Article
| Open Access Replication dynamics of recombination-dependent replication forks
Replication dynamics of recombination-dependent replication forks
Replication forks that are stalled at obstacles on the DNA template can be restarted by homologous recombination. Here, the authors show replication dynamics during homologous recombination-dependent replication fork restart by combining polymerase usage sequencing and a Monte Carlo mathematical model.
- Karel Naiman
- , Eduard Campillo-Funollet
- & Antony M. Carr
-
Article
| Open Access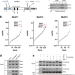 The Bloom syndrome complex senses RPA-coated single-stranded DNA to restart stalled replication forks
The Bloom syndrome complex senses RPA-coated single-stranded DNA to restart stalled replication forks
The BLM helicase interacts with the topoisomerase TOP3A and RMI1 to form the BTR complex. Here, the authors reveal that this complex contains multiple binding sites for the single-stranded DNA-binding complex RPA, and that RPA-binding stimulates BLM recruitment to stalled replication forks to promote their restart after replication stress.
- Ann-Marie K. Shorrocks
- , Samuel E. Jones
- & Andrew N. Blackford
-
Article
| Open Access High-fidelity DNA ligation enforces accurate Okazaki fragment maturation during DNA replication
High-fidelity DNA ligation enforces accurate Okazaki fragment maturation during DNA replication
DNA ligase 1 (LIG1) finalizes eukaryotic nuclear DNA synthesis by sealing Okazaki fragments using DNA end-joining reactions. Here the authors, by studying an engineered low-fidelity LIG1, reveal that LIG1 is a highly accurate DNA ligase in vivo.
- Jessica S. Williams
- , Percy P. Tumbale
- & Thomas A. Kunkel
-
Article
| Open Access Structural basis for the multi-activity factor Rad5 in replication stress tolerance
Structural basis for the multi-activity factor Rad5 in replication stress tolerance
Rad5 is a hub connecting three replication stress tolerance pathways. Here, the authors present the 3.3 Å crystal structure of a N-terminal truncated K.lactis Rad5 construct that reveals the spatial arrangement of the HIRAN, Snf2 and RING domains and structure-guided in vitro and in vivo experiments reveal multiple activities of the yeast Rad5 HIRAN domain among them a role in binding PCNA and supporting its ubiquitination.
- Miaomiao Shen
- , Nalini Dhingra
- & Song Xiang
-
Article
| Open Access Cohesin depleted cells rebuild functional nuclear compartments after endomitosis
Cohesin depleted cells rebuild functional nuclear compartments after endomitosis
The role of cohesin in organizing a functional nuclear architecture remains poorly understood. Here the authors show that cohesin depleted cells pass through endomitosis forming a multilobulated nucleus able to proceed through S-phase with typical features of active and inactive nuclear compartments and spatio-temporal patterns of replication domains.
- Marion Cremer
- , Katharina Brandstetter
- & Thomas Cremer

 Pathogenic variants in SLF2 and SMC5 cause segmented chromosomes and mosaic variegated hyperploidy
Pathogenic variants in SLF2 and SMC5 cause segmented chromosomes and mosaic variegated hyperploidy