Abstract
MicroRNAs (miRNAs) present frequently altered expression in urologic cancers including prostate, bladder and kidney cancer. The altered expression of miR-223 has been reported in cancers and other diseases in recent researches. MiR-223 is up-regulated in systemic lupus erythematosus and rheumatoid arthritis. In neoplastic diseases, miR-223 is proved to be up-expressed in plasma or serum and cancer tissues compared with normal tissues in pancreatic cancer, gastric cancer, et al. However, whether altered expression of miR-223 is associated with prostate cancer (PCa) and what it is potential functions in PCa remained unveiled. In this study, we firstly found miR-223-3p were up-regulated in prostate cancer tissues and then we study functional role of miR-223-3p in PCa using DU145, PC3 and LNCaP cell lines. Our data suggested that miR-223-3p might target gene SEPT6 and promoted the biological behavior of prostate cancer. Notably, we found increasing SEPT6 expression might reverse the biological activity induced by miR-223-3p, which might be a potential therapeutic target for PCa.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) is one of the most common malignancies in men and contributes to the second most frequent cause of cancer-associated death1. Patients with advanced or metastatic prostate cancers have a less average survival time and suffer from cancer metastasis-related symptoms. To control and well-treat PCa, methods of early cancer detection and delaying cancer progression enable patients improving survivals and their quality of life. Many researchers have found potential prognostic factors and tumor aggressiveness of PCa, including role of genetic mutation related biological pathways2,3,4,5. Like other solid cancers, PCa metastasis proceeds through a complicated series of molecular events, including angiogenesis at the site of the original tumor, local migration within the primary site, intravasation into the blood stream, survival within the circulation, extravasation of the tumor cells to the target organ and colonization of those cells within the new site2.
MicroRNAs (miRNAs, miRs) are a class of small non-coding RNAs that negatively regulate gene expression primarily through post-transcriptional modification. MiRNAs present commonly altered expression in urologic cancers including prostate, bladder and kidney cancer. They appear to be important modulators of these tumors and many of them are functionally implicated in their pathogenesis4,6,7,8,9. These miRNAs are up or down expressed and alter the prostate cancer pathways such as cell apoptosis, cell cycle control, proliferation, androgen independence and drug resistence by targeting their “seed” regions that bind complementary sequences within messenger RNA (mRNA) tails (3′ untranslated region)4. The translational applications about miRNAs include their use as disease biomarkers, in prognostic prediction and as novel treatments. The most exciting application enables potentially disease-specific individualized therapeutic targeting. The altered expression of miR-223 has been reported in cancers and other diseases in recent researches. MiR-223 is up-regulated in systemic lupus erythematosus and rheumatoid arthritis10. In neoplastic diseases, miR-223 is proved to be up-expressed in plasma or serum and cancer tissues compared with normal tissues in pancreatic cancer11, gastric cancer12,13,14, et al. However, whether altered expression of miR-223 is associated with PCa and what it is potential functions in PCa remained unveiled. In this study, we firstly found overexpression of miR-223-3p (MIMAT0000280) in PCa tissues and PCa cell lines. Our data suggested that miR-223-3p promote the biological behavior of PCa cell lines, including decreasing cell apoptosis and increasing cell migration and invasion. Future more, we found reverse regulation effects of miR-223-3p on gene SEPT6 (NM-015129) and confirmed SEPT6 was a target gene of miR-223-3p with luciferase reporter assay. The role of SEPT6 on PCa the biological activity targeted by miR-223-3p was studied and the results indicated miR-223-3p's potential role as a therapeutic target for PCa.
Results
Expression of miR-223-3p was up-regulated in PCa tissues and PCa cell lines
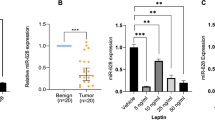
Paired 10 PCa tissues and adjacent non-cancer tissues were used to detect expression of miR-223-3p. The expression of miR-223-3p in PCa tissues was found about 1.9 fold more than the paired adjacent non-cancer tissues (2.98 ± 1.45 vs 1.55 ± 0.38). The expression level of miR-223-3p was significantly increased in PCa tissues compared with adjacent non-cancer tissues (p < 0.01) (Figure 1A). The mean Gleason score of the total 10 patients was 6.20 ± 1.16, ranging from 3 to 8. PCa samples were divided into two groups by Gleason score, group of well differentiation (n = 5, Gleason score ≤6, WD) and group of poor differentiation (n = 5, Gleason score ≥7, PD). Expression of miR-223-3p in group of PD was found significant higher than that in group of WD (4.10 ± 1.14 vs 1.86 ± 0.55, p = 0.004) (Figure 1B). Expression of miR-223-3p was also examined in different PCa cell lines LNCaP, PC3 and DU145. The results showed significant differences were discovered in each pairwise groups. All PCa cells presented significantly higher than that in non-tumor human prostate epithelial cells (RWPE-1) (all p < 0.01) (Figure 1C). In details, among PCa cell lines, expression of miR-223-3p in DU145 was in highest level and the middle level one was in PC3, while in LNCaP, the lowest level was obtained.
Expression of miR-223-3p is up-regulated in PCa tissues and PCa cell lines.
Figure 1A: The expression level of miR-223-3p was significantly increased in PCa tissues compared with adjacent non-cancer tissues (p < 0.01). Figure 1B: PCa samples were divided into two groups by Gleason score, group of well differentiation (WD) and group of poor differentiation (PD) and expression of miR-223-3p in group of poor differentiation was found significant high than that in group of well differentiation (p = 0.004). Figure 1C: Expression of miR-223-3p was examined in different PCa cell lines LNCaP, PC3 and DU145. The results showed pairwise significant differences were discovered among them, all significantly higher than that in non-tumor human prostate epithelial cells (RWPE-1) (all p < 0.01). ** on behalf of p < 0.01.
MiR-223-3p regulates biological behavior in PCa cell lines
Detection of transfection efficiency
Three PCa cell lines DU145, PC3 and LNCaP cells were divided into four groups. According to previous study, miRNA's mimics and miRNA's antisense oligonucleotides were used for alternating miRNA's expression15. To promote miR-223-3p expression, cells were transfected by miR-mimics (Sense: 5′-ugucaguuugucaaauacccca-3′; antisense:5′-ugggguauuugacaaacugaca-3′) (miR-MM). To inhibit expression of miR-223-3p, cells were knocked down by the miR-223-3p antisense oligonucleotides (5′- ugggguauuugacaaacugaca-3′) (miR-AO). Cells transfected with miRNA negative control (miR-NC) and conventional culture cells (CC-cells) were used as two control groups. Detection of the above transfections was performed on light and fluorescence microscope and by quantitative real-time polymerase chain reaction (qRT-PCR). The results of fluorescence microscope indicated transfection efficiencies in all the two groups of miR-MM and miR-AO were both more than 90% (Figure 2A1 and 2A2). QRT-PCR was carried out for miR-223-3p expression evaluation after the different transfections (Figure 2B). The results suggested the different expression levels of miR-223-3p were existed among four groups in each cell lines. MiR-223-3p contents in miR-MM groups were significantly higher than the other three groups (all p < 0.01). While expression in groups of miR-AO were lowest (all p < 0.01), leaving groups of CC-cells and miR-NC in the middle level and with no significant differences found between them. These results of fluorescence microscope and qRT-PCR demonstrated the above transfections were successful and effective.
MiR-223-3p regulates biological behavior in PCa cell lines.
Figure 2A1 and 2A2: Fluorescence microscope is used for detecting transfection efficiency for miR-MM and miR-AO transfection and the results suggested transfection efficiencies are all more than 90%. Figure 2B: MiR-223-3p expression is evaluated in groups after the different transfections, resulting in different expression levels of miR-223-3p are existed among four groups in each cell lines. MiR-223-3p contents in miR-MM groups are significantly higher than the other three groups (all p < 0.01), while expression in groups of miR-AO are lowest (all p < 0.01). Figure 2C1, 2C2 and 2C3: cell proliferation assays for DU145, PC3 and LNCaP, show that knocking down miR-223-3p expression (groups of miR-AO) leads to lowest cell proliferation, cells with up-regulated expression of miR-223-3p after miR-MM transfected present highest cell proliferation. Figure 2D, 2E1, 2E2 and 2E3: cell cycle assays for DU145, PC3 and LNCaP. Knocking down miR-223-3p expression makes cells arrest in G0/G1 sub phase and induces their apoptosis, with the proliferation index (PI) significantly lower than other groups. Figure 2F and 2G: Cell apoptosis assay. The results indicat cells with miR-223-3p knocked down present highest level of apoptosis, while cells with up-regulated expression of miR-223-3p after miR-MM transfected present lowest apoptosis (all p < 0.01). Figure 2H and 2I: In cell invasion assay, cells showed lowest invasive ability in groups of miR-AO, while in contrast, cells with up-regulated expression of miR-223-3p after miR-MM transfected present highest invasive ability (all p < 0.05).** on behalf of p < 0.01. * on behalf of p < 0.05.
Detection of biological behaviors
To figure out function of higher miR-223-3p expression in PCa cell lines, we examined the potential role of miR-223-3p in cell proliferation, apoptosis, cell cycle and invasion assays after cells were divided into the above four groups. The results of cell proliferation and cell cycle assays showed that knocking down miR-223-3p expression (groups of miR-AO) leaded to lowest cell proliferation (Figure 2C1, 2C2 and 2C3), made cells arrest in G0/G1 sub phase (Figure 2D, 2E1, 2E2 and 2E3) and induced their apoptosis, with the proliferation index (PI) significantly lower than other three groups. Increased cell apoptosis was demonstrated by cell apoptosis assay and the results indicated cells with miR-223-3p knocked down presented highest level of apoptosis (all p < 0.01) (Figure 2F and 2G). Similarly results were seen in cell invasion assay and cells showed lowest invasive ability in groups of miR-AO (all p < 0.05) (Figure 2H and 2I). While in contrast, cells with up-regulated expression of miR-223-3p after miR-MM transfected presented an opposite above biological behaviors (highest cell proliferation, lowest apoptosis and highest invasive ability). No significant differences were found between groups of CC-cells and miR-NC in the four assays (all p > 0.05).
MiR-223-3p regulated SEPT6 expression by directly targeting its 3′ UTR and promoted biological behaviors of PCa cells
To find target genes of miR-223-3p, we searched several commonly used target gene prediction websites like Targetspy, Pictar, Microrna and Mirbase. All the results suggested that 3′-UTR SEPT6 mRNA contains two highly conserved binding sites from positions 55 to 76 and 1135 to 1156 for miR-223-3p (Figure 3A), indicating SEPT6 might be a target gene of miR-223-3p. To further demonstrate this possibility, SEPT6 protein and mRNA were detected and luciferase reporter assay was performed.
MiR-223-3p regulates SEPT6 expression by directly targeting its 3′ UTR.
Figure 3A: 3′-UTR SEPT6 mRNA contains two highly conserved binding sites from position 55 to 76 and 1135 to 1156 for miR-223-3p. Figure 3B1, 3B2, 3C1 and 3C2: expression of SEPT6 mRNA and protein in PCa tissues and cell lines. Compared with adjacent non-tumor tissues and RWPE-1, lower expressions of SEPT6 mRNA and protein were found in PCa tissues and cell lines than those in adjacent non-tumor tissues and RWPE-1 (all p < 0.01). Figure 3D: Luciferase reporter assay was carried out and its result confirms that SEPT6 is directly targeted by miR-223-3p. In DU145 cells transfected by miR-MM group, a significant decreased expression of SEPT6 is observed, with about 79.8% down-regulation rate; conversely, in DU145 cells with miR-AO transfected, a significant increased expression of it was obtained, about 103.6% up-regulation rate (all p < 0.01). ** on behalf of p < 0.01.
We detected expression of SEPT6 protein and mRNA in PCa tissues and cell lines. Compared with adjacent non-tumor tissues and RWPE-1, lower expressions of SEPT6 mRNA and protein were found in PCa tissues and cell lines (all p < 0.01) (Figure 3B1, 3B2, 3C1 and 3C2). Combined with the higher miR-223-3p expression in PCa tissues and cell lines in above section, the hypothesis was established that a potential reverse regulation might be existed between miR-223-3p and SEPT6. Furthermore, we found expressions of SEPT6 mRNA and protein were lowest in DU145 while miR-223-3p expressed highest in it, thus DU145 cell line was selected to do further studies, in order to confirm the SEPT6 might be a target gene of miR-223-3p and to figure out its potential role in PCa cells.
Luciferase reporter assay was carried out and its result confirmed that SEPT6 was directly targeted by miR-223-3p. In group of DU145 cells transfected by miR-MM, a significant decreased expression of SEPT6 was observed, with about 79.8% down-regulation rate; conversely, in DU145 cells with miR-AO transfected, a significant increased expression of it was obtained, about 103.6% up-regulation rate (Figure 3D). At this point, we were sure about SEPT6 was a target gene of miR-223-3p.
Further more studies were done to interpret SEPT6's role in DU145 cell apoptosis, migration and invasion that induced by highly expressed miR-223-3p. SiRNA was used to transfect DU145 cells to regulate SEPT6 mRNA and protein expression. Three siRNAs of SEPT6 were transfected into DU145 cells and their gene silencing effects were evaluated, siRNA-SEPT6-163 (sense: 5′-AGAGAAAGUGUUACACUUCTT-3′, antisense: 5′-GAAGUGUAACACUUUCUCUTT-3′, target sequence on SEPT6: 5′- ccgaagtgtaacactttctcttt-3′, siR-S-163), siRNA-SEPT6-1219 (sense: 5′- UCAUAUGUCUCCUGUAAACTT-3′, antisense: 5′- GUUUACAGGAGACAUAUGATT-3′, target sequence on SEPT6: 5′- cagtttacaggagacatatgagg-3′, siR-S-1219) and siRNA-SEPT6-2289 (sense: 5′-UUGUUUAAGCGUGAAUUUCTT-3′, antisense: 5′- GAAAUUCACGCUUAAACAATT-3′, target sequence on SEPT6: 5′- cagaaattcacgcttaaacaacc-3′, siR-S-2289). Scrambled siRNA as negative control (sense: 5′-UUCUCCGAACGUGUCACGUTT, antisense: 5′-ACGUGACACGUUCGGAGAATT-3′, siR-S-NC) and CC-cells were used as control groups. All transfection efficiencies were detected on fluorescence microscope and the results showed all of them were more than 80% (Figure 4A), suggesting these transfections were effective. The results of western blot and qRT-PCR confirmed that mRNA (Figure 4B1) and protein (Figure 4B2) of SEPT6 in cells that transfected with siRNA-SEPT6-1219 expressed at the lowest level (all p < 0.01), obviously indicating that siR-S-1219 was the best gene-silencing vector. In two control groups of siR-S-NC and CC-cells, no significant difference was obtained (p > 0.05). Thus siR-S-1219 was selected to regulate SEPT6 expression in the following study.
SEPT6's role in DU145 cell apoptosis, migration and invasion.
Figure 4A: Transfection efficiencies were detected on fluorescence microscope and the result shows all of them were more than 80%. Figure 4B1 and 4B2: Western blot and qRT-PCR confirmed that mRNA and protein of SEPT6 in cells that transfected with siRNA-SEPT6-1219 expressed at the lowest level (all p < 0.01), obviously indicating that siR-S-1219 was the best gene-silencing vector, thus siR-S-1219 was selected to regulate SEPT6 expression in the following study. Figure 4C: The expression of miR-223-3p in groups of miR-NC + siR-S-NC and miR-NC + siR-S-1219 are higher than in the other two groups transfected with miR-AO + siR-S-1219 and miR-AO + siR-S-NC (all p < 0.01), while no significant difference is found between two groups with miR-NC or two groups with miR-AO. Figure 4D1 and 4D2: Highest expression of SEPT6 mRNA and protein are achieved in group miR-AO + siR-S-NC and lowest express of them are found in group miR-NC + siR-S-1219 (all p < 0.01). In group of miR-AO + siR-S-NC where SEPT6 mRNA and protein expressed in highest level, cells presented highest rate of cell apoptosis (Figure 4E) and lowest cell abilities of migration (Figure 4F) and invasion (Figure 4G) among four groups (all p < 0.05). On the contrary, in group of miR-NC + siR-S-1219 with lowest expression of SEPT6, cells presented lowest rate of cell apoptosis and highest cell abilities of migration and invasion (all p < 0.05). ** on behalf of p < 0.01.
To decrease expression of SEPT6 and unchange expression of miR-223-3p, DU145 was transfected with miR-NC + siR-S-1219. To increase expression of SEPT6, DU145 was transfected with miR-AO + siR-S-NC. MiR-NC + siR-S-NC and miR-AO + siR-S-1219 tranfected cells were used as control groups. The expression of miR-223-3p in groups of miR-NC + siR-S-NC and miR-NC + siR-S-1219 were higher than in the other two groups transfected with miR-AO + siR-S-1219 and miR-AO + siR-S-NC (all p < 0.01). While no significant difference was found between two groups with miR-NC or two groups with miR-AO (Figure 4C). Together with that we achieved highest expression of SEPT6 mRNA and protein in group miR-AO + siR-S-NC and lowest express of it in group miR-NC + siR-S-1219 (all p < 0.01) (Figure 4D1 and 4D2), the interference of SEPT6 expression was considered effective. Cell apoptosis, migration and invasion assays were performed subsequently. In group of miR-AO + siR-S-NC where SEPT6 mRNA and protein expressed in highest level, cells presented highest rate of cell apoptosis (Figure 4E) and lowest cell abilities of migration (Figure 4F) and invasion (Figure 4G) among four groups (all p < 0.05). On the contrary, in group of miR-NC + siR-S-1219 with lowest expression of SEPT6, cells presented lowest rate of cell apoptosis and highest cell abilities of migration and invasion (all p < 0.05).
Discussion
The altered expression of miR-223 has been found in cancers and other diseases in recent studies. MiR-223 is found up-regulated in systemic lupus erythematosus and rheumatoid arthritis10. In neoplastic diseases, miR-223 is proved to be up-expressed in plasma or serum and cancer tissues compared with normal tissues in pancreatic cancer11, gastric cancer12,13,14, esophageal squamous cell carcinoma16, hepatocellular carcinoma17,18, breast cancer19, lung cancer20, endometrioid endometrial cancer21. However, it is also found down-regulated in chronic lymphocytic leukemia22,23, nasopharyngeal carcinoma24, osteosarcoma25, intrahepatic cholangiocarcinoma26. In a study, 16 miRNAs are identified differentially expressed between acute lymphoblastic leukemia (ALL) and acute myelocytic leukemia (AML) samples, the results suggest nine are expressed at a significantly higher level in ALL than in AML, while in contrast; miR-223 and the other six miRs are expressed at a significantly higher level in AML compared to ALL27. Moreover, in previous studies, altered expression of miR-223 is demonstrated in urologic cancers, such as up-regulated in bladder cancer28. In our study, we firstly found miR-223-3p was significantly up-regulated in PCa tissues compared with the normal adjacent tissues (Figure 1A) and PCa cell lines (Figure 1C), indicating that miR-223-3p might be associated with PCa.
MiR-223 may be a predictor of cancer reoccurrence and metastasis. Compared with non-recurrent hepatocellular carcinoma, miR-223 is expressed much higher than in recurrent hepatocellular carcinoma29. In addition, highly expressed miR-223-3p is significantly associated with a higher risk for progression in adenocarcinoma patients30. Plasma levels of miR-223 increased when metastasis manifested in patients with uveal melanoma31. In recurrent ovarian cancer, miR-223 is also proved to be of potential importance as biomarkers32. In our study, miR-223-3p was proved positive correlated with Gleason scores (Figure 1B). Further study found over-expressed miR-223-3p promoted PCa cell lines proliferation (Figure 2C1, 2C2 and 2C3), leaded to less cell apoptosis (Figure 2F and 2G) and accelerated invasive ability (Figure 2H and 2I). On the contrary, knocking down miR-223-3p made cell arrest in G0/G1 sub phase (Figure 2D, 2E1, 2E2 and 2E3), induced its apoptosis and decreased DU145 cell invasive ability. These results predicted that highly expressed miR-223-3p might be associated with the onset and progression of PCa.
With base-pairing reactions, the 5′end of miRNAs include the targeting “seed” regions that can bind complementary sequences within mRNAs 3′ untranslated tails. This kind of affinity bond depends on the sequence and number of complementary seeds33. The miRNA–mRNA pair recruits a silencing complex to modulate mRNA expression. Although it is now appreciated that miRNAs are involved in a broad range of biological processes, an exploration of a miRNA downstream target genes may reveal more fully the molecular mechanisms of its role in cancers. In the previous studies, many of target genes of miR-223 have been found, such as tumor suppressor gene EPB41L313, NFI-A34, stathmin135, caprin-136, ect225, IGF-1R and CDK237. In our study SEPT6 was identified as direct target gene of miR-223-3p using qRT-PCR, western blot analysis and luciferase reporter assay (Figure 3).
Septins are filament-forming GTP-binding proteins that act as scaffolds in diverse cell functions including division, polarity and membrane remodeling. Human septins comprise a family of 13 genes that encode for more than 30 protein isoforms with ubiquitous and tissue-specific expressions. Different expression and function of septins present in multiple systems, including the cardiovascular, immune, nervous, urinary, digestive, respiratory, endocrine, reproductive and integumentary organ systems38. Septins may also belong to a kind of cancer critical genes. More septins might act as potential oncogenes or tumor suppressor genes in cancer development39. In a recent review article, the role of septins in tumorigenesis, emphasizing their significance and supporting their use as potential biomarkers in various cancer types are clearly summarized40. SEPT6 also known as SEP2 and KIAA0128, located at Xq2441, is expressed ubiquitously, plays a role in actin dynamics, cell shape and microtubule regulation and is associated with bipolar disorder, Down's syndrome, infection (bacterial, viral), schizophrenia and cancers, including skin, leukaemia and lymphoma42. In our study, SEPT6 was firstly discovered targeted by miR-223-3p and associated with PCa. The results of luciferase reporter assay presented that SEPT6 expression was significant decreased in miR-MM group, with down-regulation rate about 79.8%; conversely, it was significant increased in miR-AO group, with the up-regulation rate about 103.6% (Figure 3D). Further study found that unregulated expression of SEPT6 could inhibit the biologic activity of DU145. The results indicated that in group of miR-AO + siR-S-NC, highest expression of SEPT6 (Figure 4D1 and 4D2), leaded to highest rate of cell apoptosis (Figure 4E), lowest cell migration (Figure 4F) and invasion (Figure 4G). Converse conclusions were obtained in group of miR-NC + siR-S-1219 in which expression level of SEPT6 was lowest (Figure 4D1, 4D2, 4E, 4F and 4G). Collectively, these results might indicate that increasing expression of SEPT6 might reverse the biologic activity of PCa cells induced by miR-223-3p, which might apply a potential therapeutic target for PCa treatment.
Members of septins are proved having a tumor suppressor function and or oncogenic function40. Nevertheless, in our study, as a member of septins, SEPT6 might present as carrying tumor suppressor properties in PCa, though its exact role remained unknown. One possible mechanism might be a gene fusion and or gene interaction happened between SEPT6 and other PCa related genes. As firstly, In hematological malignancies, members of the septin family were shown to fuse to mixed-lineage leukemia (MLL), including SEPT643,44. A gene fusion is generated by a chromosomal translocation and is a frequent event in leukemias45. Secondly, septins have also been found to present altered expression or cellular localization in hormonally regulated tumors such as prostate (SEPT9)46. The interaction of overexpressed SEPT9_v1 with hypoxia-inducible factor-1a (HIF-1a) in prostate cancer results in increased cell proliferation and tumor angiogenesis, whereas the knockdown of SEPT9_v1 is associated with an increase in HIF-1a degradation, a decrease in HIF-1 transcriptional activity, cell proliferation and angiogenesis46. Another possibility is that the main mechanism by which altered gene expression leads to tumorigenesis is through septin downregulation or deletion, resulting in impairment of cell division leading to polyploidy or aneuploidy40.
There are limitations about our study. In future study, large sample of case-control study and experimental animal models are needed to analyze the role of miR-223-3p and SEPT6 in PCa's occurrence, development and prognosis. The startup mechanism of high expression of miR-223-3p and downstream pathways associated with SEPT6 in PCa should also be further explored.
In conclusion, our data supply the convince evidence of miR-223-3p up-regulation and its potential role in PCa. Notably, we found SEPT6 is a target gene of miR-223-3p and it may reverse the biological activity induced by miR-223-3p, which might supply a potential therapeutic target for PCa.
Methods
Ethics Statement
The approval for this study was obtained from the Institutional Review Board of the Second Xiangya Hospital, Central South University. Informed consent was obtained from all patients or their families for their related tissues were used. The study did not contain any identifying information about any individual subjects. All the data was kept by the administrator of the study team in a confidential manner and was not used by any other purposes. We confirm that all experiments were performed in accordance with relevant guidelines and regulations.
Tissue samples
In order to assess the expression level of miR-223-3p in prostate cancer compared with normal prostate tissue, human prostate cancer tissues and the matched adjacent non-cancer tissues were harvested from prostate cancer patients in Department of Urology, the Second Xiangya Hospital, Central South University. All samples were stored at -80°C in liquid nitrogen immediately after surgery.
Cell culture and transfection
Three prostate cell lines PC3, DU145 and LNCaP and RWPE-1 were cultured in RPMI-1640 (Sigma, St. Louis, MO, USA), with 10% FBS and 5% CO2 at 37°C. PCa cells were cultured after 24 hours and were divided into different groups. The transfections were performed on Lipofectamine™ 2000 complexes (Invitrogen, CA, USA). After 24 or 48 hours incubation in the incubator, cells were detected on light and fluorescence microscope.
QRT-PCR and Real-time PCR analysis of mRNA
Total miRNAs were purificated from human tissues and cell lines with miRNeasy Mini Kit (Qiagen #217004, USA), according to the manufacturer' instructions. QRT-PCR was carried out on the ABI 7900 system (Applied Biosystems, USA). MiR-223-3p was detected using miRNA Q-PCR Detection Kit (GeneCopoeia #R0101L, USA). Total RNA for Real-time PCR was reverse transcribed using RevertAid™ H Minus First Strand cDNA Synthesis Kit (Fermentas #K1631). All experiments were repeated three times. Standard curves were generated. The results were normalized according to internal controls. The data were presented based on the 2−ΔΔCt method47.
Western blot
Proteins were extracted at 24 or 48 hours post-transfection and used for western blot analysis. Protein lysates were separated by 10% SDS-PAGE (Sigma, St. Louis, MO, USA), then transferred onto nitrocellulose blotting membranes (Pierce, Rockford, IL, USA). Membranes were blocked and incubated with SEPT6 antibody (1:400 Proteintech,#12805-1-AP,USA) and then Goat Anti Rabbit IgG/HRP (1:5000) overnight at 4°C, detected and then visualized. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as equal protein loading control.
Cell proliferation assay
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) (Sigma, USA) assay was used for examining cell proliferation. 100ul cells (1 × 104 cells) were added into each 96-well plates. After cultured in a incubator chamber at 37°C and 5% CO2, cells were transfected with miR-223-3p mimics, miR-223-3p antisense oligonucleotides or miR-NC. MTT was diluted and 50 ul 1× MTT was dropped in to each plate after the transfected cells cultured 48 hours. The cells were gestated for another four hours. Removal out of the culture supernatant, dimethyl sulfoxide (DMSO, Sigma, USA) was added for 10 minutes. A microplate reader (Molecular Devices, Sunnyvale, USA) was performed to measure the absorbance of each plate at 570 nm.
Cell apoptosis assay
Annexin V-FITC/Propidium Iodide (PI) was purchased from Mbchem™ (Mbchem M3021, USA) to evaluate cell apoptosis. The transfected cells passed through centrifugation (300 times of gravity), suspended by PBS at 4°C and then centrifugation again. 100ul labeling buffer containing 5ul annexin V-FITC was used to label the above cells. After 15 minutes cultured in the dark, 10ul PI was added into each specimen. Diluted by cold Binding Buffer dilution, the cells were detected by flow cytometry (FCM, FACSAria, BD,USA).
Cell cycle assay
Collect about 2 × 106 transfected cells and wash them with PBS at 1000rpm for five minutes. Fixed by pre-cooling 75% ethyl alcohol, the cells was incubated overnight at 4°C. Removal the alcohol, cells was washed with PBS again. After cells suspended in a mixture combined by 800ul PBS and 1% BSA, 100ul propidium iodide (PI) (Sigma, R40432, USA) and then 100ul 10mg/ml RNase A (Sigma, R4875,USA) were added. 30 minutes later, cell detection was performed on FCM at 488nm (EPICS-XL, BECKMAN-COULTER, USA).
Transwell chamber for invasion and migration assay
Transwell chamber was built by transwell 6-well plates (Corning, USA) covered with 60-80ul Matrigel dilution (BD, USA). Cells were washed by PBS and cultured in BSA serum-free medium. The cell concentration was adjusted to 5 × 104/ml. Cell culture medium containing 1 ml FBS was added on lower chamber, while cell suspension was planted on the upper chamber. After the cells were cultured for 24 hours at 37°C with 5% CO2, the matrigel and cells on upper chamber were removed. The cells were fixed with 95% ethyl alcohol for 15–20 minutes and then stained with hematoxylin for 10 minutes. Cell counting was performed on inverted microscope at 400 times magnification.
Luciferase reporter assay
In order to figure out miR-223-3p downstream target gene, we searched the three microRNA targets gene prediction systems: PicTar (http://pictar.mdc-berlin.de/), microRNA (http://www.microrna.org/), MiRBase (http://www.mirbase.org/) and TargetSpy (http://www.targetspy.org/). The 3′UTR of SEPT6 mRNA fragment was found containing miR-223-3p responsive elements and was designed as a candidate target gene of miR-223-3p. DU145 cell line was selected for transfection and target gene detection. Lipofectamine™ 2000 (Invitrogen, CA, USA) was used for cell transfection. MIR/report gene plasmid-Lipo2000 complexes containing fragment of miR-MM, miR-AO or miR-NC were prepared and added into DU145 culture medium. The cells were cultured in 48-well plates for 48 hours at 37°C with 5% CO2. Dual-Luciferase® Reporter Assay System(Promega,DLR™,E1960, USA)was performed according to the manual instructions. Renilla luciferase intensity was used as internal control to normalize the luminescence intensity of Firefly luciferase.
Statistical analysis
All experiments were repeated three times and data was presented as mean (SD). Student's t test, one-way ANOVA and subsequent LSD test were used for for statistical analyses. Significant difference was obtained when a p value < 0.05.
References
Shaikhibrahim, Z. et al. Identification of immunity-related genes in prostate cancer and potential role of the ETS family of transcription factors in their regulation. Int J Mol Med 28, 799–807 (2011).
Hudson, B. D., Kulp, K. S. & Loots, G. G. Prostate cancer invasion and metastasis: insights from mining genomic data. Brief Funct Genomics 12, 397–410 (2013).
Menegaux, F. et al. Epidemiological study of prostate cancer (EPICAP): a population-based case-control study in France. BMC Cancer 14, 106 (2014).
Catto, J. W. et al. MicroRNA in prostate, bladder and kidney cancer: a systematic review. Eur Urol 59, 671–81 (2011).
He, H. C. et al. MicroRNA-23b downregulates peroxiredoxin III in human prostate cancer. FEBS Lett 586, 2451–8 (2012).
Mlcochova, H., Hezova, R., Stanik, M. & Slaby, O. Urine microRNAs as potential noninvasive biomarkers in urologic cancers. Urol Oncol 32, 41.e1–9 (2014).
Fendler, A., Stephan, C., Yousef, G. M. & Jung, K. MicroRNAs as regulators of signal transduction in urological tumors. Clin Chem 57, 954–68 (2011).
Schaefer, A. et al. MicroRNAs and cancer: current state and future perspectives in urologic oncology. Urol Oncol 28, 4–13 (2010).
Coppola, V., De Maria, R. & Bonci, D. MicroRNAs and prostate cancer. Endocr Relat Cancer 17, F1–17 (2010).
Wang, H., Peng, W., Ouyang, X., Li, W. & Dai, Y. Circulating microRNAs as candidate biomarkers in patients with systemic lupus erythematosus. Transl Res 160, 198–206 (2012).
Bloomston, M. et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 297, 1901–8 (2007).
Petrocca, F. et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13, 272–86 (2008).
Li, X. et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res 9, 824–33 (2011).
Li, B. S. et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One 7, e41629 (2012).
Jia, Z., Wang, K., Wang, G., Zhang, A. & Pu, P. MiR-30a-5p antisense oligonucleotide suppresses glioma cell growth by targeting SEPT7. PLoS One 8, e55008 (2013).
Zhang, C. et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem 56, 1871–9 (2010).
Qi, P. et al. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One 6, e28486 (2011).
Zhou, J. et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol 29, 4781–8 (2011).
Yang, M. et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer 10, 117 (2011).
Chen, X. et al. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer 130, 1620–8 (2012).
Jia, W. et al. Identification of four serum microRNAs from a genome-wide serum microRNA expression profile as potential non-invasive biomarkers for endometrioid endometrial cancer. Oncol Lett 6, 261–267 (2013).
Calin, G. A. et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353, 1793–801 (2005).
Fazi, F. et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell 12, 457–66 (2007).
Zeng, X. et al. Circulating miR-17, miR-20a, miR-29c and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS One 7, e46367 (2012).
Xu, J. et al. MiR-223/Ect2/p21 signaling regulates osteosarcoma cell cycle progression and proliferation. Biomed Pharmacother 67, 381–6 (2013).
Karakatsanis, A. et al. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222 and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog 52, 297–303 (2013).
Wang, Y. et al. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis 44, 191–7 (2010).
Gottardo, F. et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol 25, 387–92 (2007).
Han, Z. B. et al. Identification of recurrence-related microRNAs in hepatocellular carcinoma following liver transplantation. Mol Oncol 6, 445–57 (2012).
Sanfiorenzo, C. et al. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One 8, e54596 (2013).
Achberger, S. et al. Circulating immune cell and microRNA in patients with uveal melanoma developing metastatic disease. Mol Immunol 58, 182–6 (2014).
Laios, A. et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer 7, 35 (2008).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–33 (2009).
Zardo, G. et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood 119, 4034–46 (2012).
Wong, Q. W. et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology 135, 257–69 (2008).
Gong, B. et al. Caprin-1 is a novel microRNA-223 target for regulating the proliferation and invasion of human breast cancer cells. Biomed Pharmacother 67, 629–36 (2013).
Nian, W. et al. miR-223 functions as a potent tumor suppressor of the Lewis lung carcinoma cell line by targeting insulin-like growth factor-1 receptor and cyclin-dependent kinase 2. Oncol Lett 6, 359–366 (2013).
Dolat, L., Hu, Q. & Spiliotis, E. T. Septin functions in organ system physiology and pathology. Biol Chem 395, 123–41 (2014).
Liu, M., Shen, S., Chen, F., Yu, W. & Yu, L. Linking the septin expression with carcinogenesis. Mol Biol Rep 37, 3601–8 (2010).
Connolly, D., Abdesselam, I., Verdier-Pinard, P. & Montagna, C. Septin roles in tumorigenesis. Biol Chem 392, 725–38 (2011).
Peterson, E. A. & Petty, E. M. Conquering the complex world of human septins: implications for health and disease. Clin Genet 77, 511–24 (2010).
Mostowy, S. & Cossart, P. Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol 13, 183–94 (2012).
Megonigal, M. D. et al. t(11;22)(q23;q11.2) In acute myeloid leukemia of infant twins fuses MLL with hCDCrel, a cell division cycle gene in the genomic region of deletion in DiGeorge and velocardiofacial syndromes. Proc Natl Acad Sci U S A 95, 6413–8 (1998).
Osaka, M., Rowley, J. D. & Zeleznik-Le, N. J. MSF (MLL septin-like fusion), a fusion partner gene of MLL, in a therapy-related acute myeloid leukemia with a t(11;17)(q23;q25). Proc Natl Acad Sci U S A 96, 6428–33 (1999).
Look, A. T. Oncogenic transcription factors in the human acute leukemias. Science 278, 1059–64 (1997).
Amir, S., Golan, M. & Mabjeesh, N. J. Targeted knockdown of SEPT9_v1 inhibits tumor growth and angiogenesis of human prostate cancer cells concomitant with disruption of hypoxia-inducible factor-1 pathway. Mol Cancer Res 8, 643–52 (2010).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–8 (2001).
Acknowledgements
This work is supported by the Fundamental Research Funds for the Central Universities of Central South University in 2013 (no. 2013zzts095) and China Hunan Provincial Science & Technology Department in 2013 (no. 2013TT2002).
Author information
Authors and Affiliations
Contributions
Y. Wei and J.Y. conceived the idea and concept. Y.Wang, Z.L. and Z.Z. designed the experiments. L.Y., Z.D., S.O. and H.W. analyzed the data. Y.Wei, Z.Y., K.Z., Y.G., B.Y. and Z.W. performed the experiments. Y.Wei and J.Y. wrote the manuscript. All authors reviewed the manuscript before submission.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Wei, Y., Yang, J., Yi, L. et al. MiR-223-3p targeting SEPT6 promotes the biological behavior of prostate cancer. Sci Rep 4, 7546 (2014). https://doi.org/10.1038/srep07546
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07546
This article is cited by
-
The miR-1290/OGN axis in ovarian cancer-associated fibroblasts modulates cancer cell proliferation and invasion
Journal of Ovarian Research (2024)
-
GABA induced by sleep deprivation promotes the proliferation and migration of colon tumors through miR-223-3p endogenous pathway and exosome pathway
Journal of Experimental & Clinical Cancer Research (2023)
-
miR-223-3p carried by cancer-associated fibroblast microvesicles targets SORBS1 to modulate the progression of gastric cancer
Cancer Cell International (2022)
-
Investigation of VHL gene associated with miR-223 in clear cell renal cell carcinoma
Molecular Biology Reports (2022)
-
Circular RNA circRNA_0000094 sponges microRNA-223-3p and up-regulate F-box and WD repeat domain containing 7 to restrain T cell acute lymphoblastic leukemia progression
Human Cell (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.