Abstract
Early-onset schizophrenia (EOS) offers a unique opportunity to study pathophysiological mechanisms and development of schizophrenia. Using 26 drug-naïve, first-episode EOS patients and 25 age- and gender-matched control subjects, we examined intrinsic connectivity network (ICN) deficits underlying EOS. Due to the emerging inconsistency between behavior-based psychiatric disease classification system and the underlying brain dysfunctions, we applied a fully data-driven approach to investigate whether the subjects can be grouped into highly homogeneous communities according to the characteristics of their ICNs. The resultant subject communities and the representative characteristics of ICNs were then associated with the clinical diagnosis and multivariate symptom patterns. A default mode ICN was statistically absent in EOS patients. Another frontotemporal ICN further distinguished EOS patients with predominantly negative symptoms. Connectivity patterns of this second network for the EOS patients with predominantly positive symptom were highly similar to typically developing controls. Our post-hoc functional connectivity modeling confirmed that connectivity strength in this frontotemporal circuit was significantly modulated by relative severity of positive and negative syndromes in EOS. This study presents a novel subtype discovery approach based on brain networks and proposes complex links between brain networks and symptom patterns in EOS.
Similar content being viewed by others
Introduction
Early-onset schizophrenia (EOS) is a rare and severe form of schizophrenia characterized by psychotic symptoms before the age of 18, worse prognosis than adult-onset counterparts1,2,3 and greater genetic liability4. EOS opens a unique opportunity to study the pathophysiological development and mechanisms of schizophrenia5. Neuroimaging studies have reported widespread brain structure abnormalities in EOS6,7,8, suggesting that the functional impairments are distributed across brain networks.
The relationship between EOS and impairments in large-scale functional brain networks, however, remains unclear. Which brain networks are vulnerable to schizophrenia and how dysfunctions of these networks are related to the manifestation of symptoms are open questions. There have been extensive studies on intrinsic functional connectivity networks in adult-onset schizophrenia9,10,11, but the complex reorganization of functional networks during brain maturation makes it challenging to parse pathophysiological effects from normal developmental changes in brain networks. To our knowledge, resting-state fMRI studies on drug-naïve, first-episode EOS patients are completely lacking.
In the present study, we recruited 26 first-episode, drug-naïve EOS patients and 25 matched typical developing control (TDC) teenagers to investigate the characteristics of intrinsic connectivity networks (ICNs) measured using resting-state fMRI. As pointed out by Kapur et al.12, “the current diagnostic system was not designed to facilitate biological differentiation and it does not”. Acknowledging the high potential that patients diagnosed based on behavioral symptoms may be highly heterogeneous in pathology, we decided against simply comparing ICNs between pre-defined subject groups under the assumption that all patients share homogeneous ICN characteristics. Instead, we adopted the spirit of the ‘Research Domain Criteria’ project13 and aimed to utilize the inter-subject heterogeneity of ICNs to inform potential subtypes of patients.
We employed a systematic data-mining approach, “generalized ranking and averaging independent component analysis by reproducibility” (gRAICAR14,15,16), to investigate whether the subjects can be grouped into communities according to the characteristics of their ICNs (See Figure 1 for a graphical demonstration). The subjects in a highly homogeneous community derived from an ICN could share a common pathophysiology that is different from other subjects. This homogeneous subject community thus may suggest a pathophysiological subtype. The clinical diagnosis and behavioral measures were not used to define subject groups prior to gRAICAR analysis, but they were associated to the ICN-derived subject communities to interpret the findings in neuroimaging data. gRAICAR identified 15 ICNs that showed modest to high consistency across all subjects. We further examined the subject communities revealed by each of these 15 ICNs and interpreted the ICN-derived subject communities using clinical symptom patterns. We found two ICNs whose subject community profiles exhibited significant associations with clinical diagnosis or relative severity of the positive and negative symptoms.
Demonstration of gRAICAR analysis flow.
For simplicity, we demonstrate the workflow with only three subjects (denoted as S1, S2 and S3). First, the fMRI data for the subjects are decomposed individually using spatial independent component analysis (ICA) into spatial components (ICs). The resultant component maps are presented in the light blue layer. Suppose that for each subject we obtain four ICs. The circles representing the ICs that are color coded to indicate which subject they are from. gRAICAR is applied to these ICs from individual subjects. In the pink layer, we present a distance space. The similarity between all ICs is depicted in this distance space. The aim of gRAICAR is to identify ICs that are from different subjects but are close to each other (as marked with black dashed circles). These ICs are clustered to form group-level aligned components (ACs). For each AC, according to the distance between its composing ICs, we get a similarity matrix indicating the similarity across all subjects. Based on this similarity matrix, a community detection algorithm can be applied to each AC to identify homogeneous subject communities among all subjects.
Results
The dataset included 26 drug-naïve, first-onset EOS patients and 25 TDC subjects. Demographical and clinical information of the patients and healthy controls are shown in Table 1. There was no significant difference between the two groups in terms of sex ratio (χ2(1) = 0.002, p = 0.98), age (t(49) = −0.20, p = 0.84), or head motion parameters (t(49) = −0.24, p = 0.81) as measured by the mean of frame-wise displacement.
Preprocessed functional images of all subjects were analyzed using gRAICAR. The resting-state scan for each subject was decomposed into spatial components using independent component analysis. gRAICAR then searched for similar spatial components across all subjects and clustered them as aligned components (ACs), regardless of their group identities. For each group-level AC, gRAICAR computed a similarity matrix showing inter-subject similarity between their spatial components that contributed to this AC (see Figure 1). The contribution of each subject to each AC was examined with a permutation test. A k-clique community detection algorithm was applied to the similarity matrix of each AC to identify highly homogeneous subject communities based on similarity of spatial components.
This approach identified 15 ACs representing ICNs with at least 5 significantly contributing subjects. The inter-subject similarity matrix for each ICN (e.g., Figure 2B) reflected a subject community profile (e.g., Figure 2C). The ICN-derived subject communities reflect potential subgroups of subjects that share similar ICN characteristics. These subgroups were obtained independently from clinical diagnosis. To interpret these findings, we hypothesized that the subjects could be grouped into communities according to their clinical measures that maximally agreed with the ICN-derived communities. In the following sections, we report two ICN-derived subject communities that are most related to clinical measures. The other 13 ICNs are presented in Supplementary Figure 1, though they have much weaker relationships with the clinical measures.
gRAICAR reveals the precuneus-angular gyri (PCU-AG) network is associated with EOS diagnosis.
(A) PCU-AG network rendered onto cortical surfaces of the brain. The network consists of three brain regions located in the precuneus, left angular gyrus and right angular gyrus. The maps were thresholded at |Z| > 1.5 for better visualization on the surfaces. (B) Similarity matrix of the PCU-AG networks across all subjects. Both horizontal and vertical axes represent subjects. For visualization purpose, the subjects are grouped into TDC and EOS groups and the pink solid lines mark the boundary between the two subject groups. (C) A multi-dimension scaling graph visualizing inter-subject similarity from (B) with community detection results. In the graph, each node represents a subject and the color of the node encodes the subject group (TDC or EOS, see the legend). The distance between every pair of nodes indicates the similarity between the ICNs from the two corresponding subjects. The community detection algorithm identified a number of maximal-cliques (as indicated with gray lines) and merged those with overlapping nodes, yielding the subject community marked by pink circles. 68% (17/25) of TDC subjects were included in the community, while only 34% (9 of 26) of EOS subjects were included.
An ICN associated with EOS diagnosis
One ICN formed by the precuneus (PCU; x = −9 mm, y = −69 mm, z = 45 mm in MNI space) and left and right angular gyri (l-AG; x = +48, y = −72, z = +21 and r-AG; x = −39, y = −81, z = +24) reflected a subject community profile associated with the EOS/TDC diagnosis (Figure 2). The inter-subject similarity matrix of this PCU-AG ICN (Figure 2B) depicts similarities between all of the subjects that can be visualized in a graph using a multi-dimensional scaling approximation (Figure 2C). According to the binary community profile, 68% (17 of 25) TDC subjects were included in the community, whereas 35% (9 of 26) EOS subjects were included. The mean age of the 26 subjects included in this community was 14.5 (standard deviation = 2.62) and the mean age of subjects not included in this community was 14.4 (standard deviation = 2.36). There was no significant difference in age between the subjects within and outside this community (t(49) = −0.21, p = 0.83).
To quantify the association between the ICN-derived inter-subject relationship and the clinical diagnosis, we constructed a clinical similarity matrix17 with all connections between TDC subjects as 1 and all other inter-subject connections as 0 (see Supplementary Methods “Linking ICN-derived subject communities to clinical characteristics”). This clinical similarity matrix was correlated with the ICN-derived similarity matrix from each of the 15 ICNs. The correlation coefficient for the PCU-AG ICN was r = 0.20 (p < 10−5, multiple comparison corrected), which was the largest among the 15 ICNs. The significance of this association was determined by a permutation test where the diagnosis labels (EOS vs. control) of the subjects were randomly permuted 1000 times. This observation indicates a significant association between the presence of this PCU-AG ICN and the clinical diagnosis.
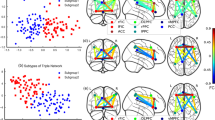
An ICN associated with multivariate symptom patterns
The second ICN's subject community profile reflected the relative severity of the positive and negative symptoms in EOS patients, as rated by the Positive and Negative Syndrome Scale (PANSS). As Figure 3A shows, this ICN consisted of left superior temporal gyrus (l-STG; x = +48, y = −24, z = −9), right superior temporal gyrus (r-STG; x = −63, y = −27, z = 0), left inferior frontal gyrus (l-IFG; x = +54, y = +18, z = −6) and right inferior frontal gyrus (r-IFG; x = −48, y = +18, z = −9). We refer to this ICN as the STG-IFG network. The subject community detected based on the inter-subject similarity matrix of this ICN contained 31 subjects. 88% of EOS patients with positive > negative scores (p > n) and 76% of TDC subjects were identified as members of this subject community, whereas only 38% of EOS patients with negative > positive scores (n > p) were included in the community (three subjects with equal positive and negative scores and two without PANSS score records were excluded from this calculation). The mean age of the 31 subjects included in the community was 14.3 (standard deviation = 1.99) and the mean age of the 15 subjects not included in the community was 14.7 (standard deviation = 2.70). There was no significant difference in age between the subjects within and outside the community (t(44) = 0.54, p = 0.59). These observations indicate that the p > n EOS patients have an ICN similar to TDC subjects, but not the n > p EOS patients. These results suggest a novel hypothesis regarding the association between this ICN and the multivariate symptom patterns in EOS patients.
gRAICAR reveals the superior temporal gyri-inferior frontal gyri (STG-IFG) network is associated with positive-negative symptom patterns.
(A) STG-lFG network rendered onto cortical surfaces of the brain. This network consists of bilateral superior temporal gyri and bilateral inferior frontal gyri. The maps were thresholded at |Z| > 1.5 for better visualization on the surfaces. (B) Similarity matrix of the STG-IFG networks across all subjects. Both horizontal and vertical axes represent subjects. For visualization purpose, the subjects are grouped into TDC, p > n EOS and n > p EOS groups. The pink solid lines mark the boundary between TDC subjects and EOS patients and the blue dashed lines indicate the boundary between p > n EOS and n > p EOS patients. (C) A multi-dimension scaling graph visualizing inter-subject similarity from (B) with community detection results. In the graph, each node represents a subject and the color of the node encodes the subject group (TDC, p > n EOS and n > p EOS, see the legend). The pink circles indicate subject memberships of the community detected based on inter-subject similarity matrix in (B). 76% (19/25) of TDC subjects and 88% (7/8) of EOS patients with positive > negative scores (p > n) were identified as members of this community, while only 38% (5/13) of EOS patients with negative > positive scores (n > p) were identified as members of the subject community.
We quantified this novel association by constructing a clinical similarity matrix that represented all TDC subjects and the EOS patients with relatively higher positive symptom scores as one community (the connections between these subjects were 1 and the others are 0). As a result, the correlation coefficient between the ICN-based similarity matrix and the clinical similarity matrix was r = 0.27 (p < 10−5, multiple comparison corrected). The significance of this association was evaluated using a permutation test where the clinical labels (EOS p > n, EOS n > p and control) were randomly permuted for 1000 times. This observation indicates that the EOS patients with more predominant positive symptoms share similarity with the TDC subjects in this STG-IFG ICN, while the EOS patients with more predominant negative symptoms are associated with abnormality in this STG-IFG network.
Reliability of the gRAICAR findings
Since the associations between the ICN-derived subject communities and the clinical measure/diagnosis-derived communities were detected based on a limited sample (n = 51), we further evaluated the reliability of detecting the associations (i.e., to rule out the possibility that the ICN-symptom associations reported above can only be found in this specific sample). As mentioned above, the ICN-symptom associations were quantified using correlation coefficients (r) between the inter-subject similarity matrices derived independently from ICNs and from clinical measures. We thus evaluated the generalizability of the associations using sample distributions of these correlation coefficients. A sample distribution of a correlation coefficient takes into account the randomness in the sampling procedure and provides an estimate of the correlation coefficients at the population level. We employed a bootstrap approach to estimate the sample distributions. The gRAICAR procedure was applied to the bootstrapped samples to compute ICN-derived inter-subject similarity matrices. The correlation coefficients between these ICN-derived similarity matrices and the clinical measure-derived similarity matrices were then calculated to generate the sample distribution (see Methods).
The sample distribution of the association between the PCU-AG ICN and the clinical diagnosis yielded a mean of r = 0.21 and a 95% confidence interval of [0.03, 0.43]. 99% of the correlation coefficients were larger than 0. The sample distribution of the association between the STG-IFG ICN and the relative positive-negative symptom severity showed a mean of r = 0.23 and a 95% confidence interval of [−0.02, 0.47]. 97% of the correlation coefficients were larger than 0. These observations indicate that the ICN-symptom associations reported above are highly likely to be discovered by gRAICAR in different datasets of the same sample size.
We also evaluated the influence of parameters in the k-clique community detection algorithms (see Methods). As a result, the subject communities reflected in the two ICNs are robust across different choices of parameters. Technical details are presented in Supplementary Methods (“Evaluating the Influence of Parameters on Community Profiles”) and Supplementary Figure 2.
Analysis of functional connectivity strength
The gRAICAR results revealed that the inter-subject similarities reflected in two ICNs are associated with clinical diagnosis or symptom patterns. We then performed post-hoc functional connectivity analyses within the PCU-AG and STG-IFG ICNs, aiming to investigate the strength of functional connectivity between the regions in the two identified ICNs. Unlike the gRAICAR analysis that attempted to associate large-scale network patterns (obtained using independent component analysis) with clinical symptoms, the following analysis focused on examining the association between the functional connectivity strength, measured using the commonly used Pearson's correlation coefficient between two regions and the symptom patterns. In addition, potential confounding factors such as age, gender and degree of head motion during the fMRI scan were controlled in the post-hoc analyses. These analyses provided supporting evidence for the findings from the gRAICAR analysis.
For the PCU-AG ICN, we obtained three regions of interest (ROIs) by applying a threshold of Z > 2.0 and a cluster size >8 voxels to its brain map, including the PCU, the l-AG and the r-AG. The time series at each voxel within these ROIs was extracted to construct a voxel-wise correlation matrix. The correlation coefficients between voxels belonging to different ROIs were converted into Fisher's Z values and then averaged to obtain a metric of inter-ROI functional connectivity (inter-FC) for each connection. A network model highlighting the functional connectivity difference between TDC and EOS subjects is shown in Figure 4A and the corresponding statistical comparisons are displayed in Figure 4B. A linear model was used to examine the differences in inter-FC between TDC and EOS subjects, where age (centered to mean) and gender (coded as a factor) were included as covariates. The results showed higher inter-FC in TDC than in EOS subjects in connections between the PCU and the r-AG (connection 1 in Figure 4A, t(46) = 2.35, p = 0.01) and between the PCU and the l-AG (connection 2 in Figure 4A, t(46) = 2.07, p = 0.02). These results were retained when the head motion parameters were also included as a covariate in the statistical tests (PCU – r-AG connection: t(45) = 2.36, p = 0.01; PCU – l-AG connection: t(45) = 2.07, p = 0.02). These observations confirmed that EOS patients exhibit aberrant functional connectivity within the PCU-AG ICN.
Correlation-based functional connectivity analyses validate gRAICAR findings.
(A) Map of the precuneus-angular gyri (PCU-AG) network showing inter-regional connections exhibiting significant (red lines) and non-significant (blue line) differences in connectivity strength (Fisher's Z) between EOS and TDC groups. (B) Bar graph showing statistical details comparing functional connectivity strength between EOS and TDC groups. Labels along the horizontal axis correspond to the connections marked on (A). The connections between the PCU and bilateral AG show significant differences between EOS and TDC groups. (C) Map of the superior temporal gyri-inferior frontal gyri (STG-IFG) network showing inter-regional connections exhibiting significant (red lines) and non-significant (blue line) differences in connectivity strength (Fisher's Z) between EOS n > p and TDC groups. (D) Bar graph showing statistical details comparing functional connectivity strength across TDC, EOS p > n and EOS n > p groups. Labels along the horizontal axis correspond to the connections numbered on (C). The connections between r-STG and r-IFG, r-STG and l-IFG and l-STG and l-IFG exhibit significant differences in connectivity strength between TDC and EOS n > p.
For the STG-IFG ICN associated with the multivariate symptom patterns, we computed the inter-FC across four ROIs (l-STG, r-STG, l-IFG and r-IFG) in the same way as described above, yielding inter-FC values for six connections. The EOS patients were separated into p > n (8 patients) and n > p (13 patients) groups based on their PANSS scores. The connections between every pair of ROIs were compared across TDC, p > n EOS and n > p EOS groups. A network model highlighting the functional connectivity difference between TDC and n > p EOS subjects is shown in Figure 4C and the corresponding statistical comparisons are displayed in Figure 4D. The functional connectivity was significantly stronger in TDC compared with n > p EOS in the connections between r-STG and r-IFG (connection 2 in Figure 4C, t(34) = 1.73, p = 0.04), r-STG and l-IFG (connection 3 in Figure 4C, t(34) = 2.35, p = 0.01) and l-STG and l-IFG (connection 5 in Figure 4C, t(34) = 1.96, p = 0.03). In contrast, p > n EOS did not show any significant differences from TDC in any connections. When comparing p > n EOS patients with n > p EOS patients, we observe significantly stronger connections in p > n EOS patients between r-STG and r-IFG (t(17) = 2.48, p = 0.01) and between r-STG and l-IFG (t(17) = 2.24, p = 0.02). These observations were retained when the head motion parameters were also included as a covariate in the comparison (in the TDC vs. n > p EOS comparison: r-STG – r-IFG: t(33) = 1.73, p = 0.04; r-STG – l-IFG: t(33) = 2.33, p = 0.01; l-STG – l-IFG: t(33) = 1.93, p = 0.03; in the p > n EOS vs. n > p EOS comparison: r-STG – r-IFG: t(16) = 2.45, p = 0.01; r-STG – l-IFG: t(16) = 2.17, p = 0.03). These results confirmed a novel association that the dysfunction of STG-IFG ICN reflects a predominance of negative symptoms in EOS patients. This ICN in EOS dominated by positive symptoms is highly similar to that in healthy population.
To further verify the above findings, we examined the interaction effect of positive and negative scores on the functional connectivity strength within this ICN. We built a regression model to fit each connection, with positive scores, negative scores, positive-negative interactions, age, gender and head motion parameters as regressors. The inter-FC of EOS patients for each connection was converted into a percentage change relative to the mean inter-FC value from all TDC subjects, which was used as a dependent variable in the models. Figure 5 shows the fitted inter-FC (colors indicate strength) for each of the six connections as a function of both positive and negative scores. The interaction between the two scores significantly affects the connection strength between the l-STG and l-IFG (connection 5 in Figures 4C and 5E, t(17) = 1.97, p = 0.03). Intuitively, the effects of the interaction between positive and negative scores on the inter-FC are reflected in the color gradient that changes predominantly along the diagonals of Figure 5E. Similar trends predicting functional connectivity strength changes along the left-to-right descending diagonal were also observed in connections r-STG – l-IFG and l-STG – r-IFG (Figure 5C–D), although they were not statistically significant. The significant interaction effects of positive-negative scores on functional connectivity strength in the STG-IFG ICN further support that the abnormality of the STG-IFG functional connectivity is relevant to the bivariate relationship between positive and negative symptoms.
Functional connectivity of the STG-IFG network is associated with multivariate symptom patterns.
Panels (A)–(F) correspond to the six inter-regional connections shown in Fig. 3C. The horizontal axis of each panel represents positive scores from PANSS and the vertical axis indicates negative scores. The black squares mark EOS n > p patients and the gray circles EOS p > n. The colors represent the difference in functional connectivity strength relative to TDC (predicted by the models), where 100% indicates the connectivity strength is the same as the mean strength of TDC subjects. The predictions were obtained from linear models including positive scores, negative scores, the interaction between the two scores and head motion parameters as regressors. The interaction between positive and negative scores exhibits a significant effect on functional connectivity strength in (E), as reflected by the changes of functional connectivity strength along the diagonal. The same trend is also shown in (C) and (D), although the interaction effect is not statistically significant.
Discussion
Uncovering characteristics of brain networks is fundamental to understanding the pathophysiological roots of early-onset schizophrenia. This task is challenging due to the lack of a priori knowledge about brain dysfunctions in early disease progression, confounding effects of psychoactive drugs and the heterogeneity in developing brains7. Moreover, it is unlikely to find a powerful one-to-one mapping between biological alteration and mental disorder that is defined based on expressed feelings and observed behavior12. These challenges motivated us to apply a systematic data-mining approach14,15,16 to investigate the characteristics of ICNs with a sample of 26 first-episode, drug-naïve EOS patients and 25 age- and sex-matched TDC subjects. We report intrinsic functional brain networks whose presence/absence in ICA results are associated with either the clinical diagnosis of EOS or the multivariate symptom patterns reflecting the balance between positive and negative symptoms in the EOS population.
The core findings of this study are novel associations between ICN characteristics and clinical symptom patterns. Specifically, we found two reliable associations: the PCU-AG ICN is associated with clinical diagnosis of EOS; the STG-IFG ICN is associated with the relative severity of the positive and negative symptoms. These findings resulted from a systematic data mining approach, gRAICAR, which pools all patients and normal controls together to detect highly homogeneous subject communities according to the similarity of their ICNs. The understudied EOS population is especially suitable for such a data mining approach that does not impose a strong assumption on group homogeneity.
Our results showed that an ICN represented by the PCU and bilateral AG could be consistently identified in TDC subjects but not in EOS patients, a result echoed by a face discrimination study showing abnormal activations in cuneus, PCU and inferior temporal cortex in EOS patients18. These observations are further supported by findings from studies on brain structure alterations in EOS1,8,19,20,21. Numerous studies have suggested that the PCU and AG are important multimodal hubs in the brain for both structural and functional connectivity22,23,24. Thus, the disruption of the PCU-AG connections we observed in EOS patients may impair the integration of and interactions between multiple brain networks, potentially leading to the disorganized cognitive and emotional functions associated with psychotic symptoms.
The PCU and AG are frequently reported as the posterior-lateral part of the default mode network25,26, which is considered to play an essential role in consciousness27 and visuospatial cognition28. Alternations in the default mode network have previously been reported to be relevant to schizophrenia29,30,31. Our results reveal default mode network dysfunction in EOS and further imply that the PCU-AG network is one of the primary targets affected by schizophrenia.
The dysfunction we observed in the PCU-AG network further indicates its central role in the neurodevelopment of schizophrenia. This argument is supported by a study on brain structure in EOS demonstrating that gray matter loss in parietal association cortex, where the PCU-AG network is located, is the beginning of a dynamic wave of gray matter loss spreading to other parts of the brain over a 5-year developmental course8. Such aberrant maturation of parietal association cortex could lead to functional abnormalities that progressively disturb the integration of information from lower-level (sensory) networks and lead to the onset of psychotic symptoms.
We identified another ICN formed by bilateral STG and bilateral IFG that was consistently detected in TDC subjects and EOS patients with more severe positive than negative symptoms, but not in EOS patients with more severe negative than positive symptoms. While most schizophrenia studies in mental disorders focus on examining correlations between brain abnormalities and positive symptoms32, our results highlight a relationship between the multivariate symptom patterns and an ICN in EOS patients. This suggests a new, more holistic perspective bridging brain dysfunction and clinical symptoms.
Our findings of the STG-IFG ICN are consistent with abnormalities in the temporal and frontal lobes of adult-onset schizophrenia patients that are frequently reported in existing literature33,34, but the relationships between these abnormities and cognitive deficits are not yet clear35,36. The abnormal STG-IFG connections we observed fit well with previous structural and functional findings on fronto-temporal connectivity in schizophrenia32,37,38,39. In a comprehensive study on brain structural and functional correlates of subclinical psychotic symptoms in children aged 11–13, Jacobson et al. reported aberrant prefrontal–temporal dysfunction in this population. Specifically, they found abnormally decreased activity in the right frontal and bilateral temporal cortex for response inhibition in the subclinical psychotic group. In their voxel-base morphometric analysis, an increase of grey matter density was observed in the STG in children with subclinical psychotic symptoms40. In addition, the same group recently reported reduced intrinsic functional connectivity between the right IFG and other regions relevant to inhibition control41. Our findings in the STG-IFG ICN echoed these previously observed abnormalities in frontal and temporal lobes in children.
Furthermore, longitudinal studies have suggested that negative symptoms develop before positive symptoms and typically before the severity of the syndrome meets the clinical diagnostic criteria for EOS42,43. Our study for the first time links this early symptom pattern (n > p) to the absence of an ICN, suggesting a potential biomarker for early detection of EOS: the absence of the STG-IFG network may predict future EOS before the psychotic symptoms reach clinical diagnosis criteria.
Functional connectivity strength analyses provide additional insights into how patterns of positive and negative symptoms modulate the strength of functional connectivity between these regions. Interestingly, we found a significant interaction effect between positive and negative scores on functional connectivity strength; EOS patients exhibiting more severe negative symptoms relative to positive symptoms had significantly weaker strength in STG-IFG connections. These observations support previous findings showing a strong relationship between negative symptoms and frontal lobe dysfunction41 and further expand this univariate theory toward a multivariate relationship.
Our findings provide evidence that the associations between clinical symptoms and brain dysfunctions are not limited to one-to-one mappings12. The positive and negative scores in the PANSS have been considered orthogonal in numerous factor analyses44,45,46. This orthogonality predicts that multivariate symptom patterns are more informative than single symptom dimensions. Our study demonstrates the power of this approach with a finding based on the multivariate symptom-brain dysfunction association in EOS, thereby providing novel hypotheses for further examination. An interesting question to be examined is whether genetic profiles of the patients can be associated with the hypothesized phenotypes.
From a methodological perspective, we presented an application of a novel neuroimaging data-mining approach, gRAICAR14,15,16, which has unique advantages in searching for potential subtypes of mental disorders according to functional neuroimaging data47. With potential disorder subtypes and complex interactions from multiple dimensions of symptoms48,49, neuroimaging data are usually heterogeneous. Findings under the assumption of group homogeneity often cannot be replicated50 and cannot serve as criteria to distinguish one disorder from another12. The data-mining approach proposed here does not assume within-group homogeneity. Instead, it characterizes the heterogeneity in a way that allows automated detection of highly homogeneous subject communities based on the data. It thus provides a neuroimaging-based exploratory tool for the big data era and is able to generate hypotheses for further examination.
There are several limitations in the current study. First, the sample size in this study (26 EOS patients, 25 controls) is not ideal in the sense of a large-scale data-mining, because the limited sample size reduced our statistical power so that we were not able to confidently propose more associations between ICN and symptom patterns, besides the two associations that our reliability analysis has validated. Collecting more data will help us to detect more subtle brain-symptom associations. Second, the interpretation of relative severity of positive and negative symptoms is not well established in previous studies. Compared with clinical symptoms, cognitive deficits tend to be more stable in schizophrenia and many studies have found their relevance to brain dysfunctions. Unfortunately, we were unable to obtain systematic cognitive measures for our subjects. In future studies, relationships between ICNs and multi-dimensional cognitive deficits should be investigated. Third, the diagnoses on the EOS patients were not reached employing a standardized interview, such as the Structured Clinical Interview for DSM-IV Axis I Disorders (SCID-I), since the SCID-I is not valid for child and adolescent population.
Methods
Participants
Thirty-two drug-naïve, first-episode EOS patients (age range: 9.0–17.9 years) and 30 TDC subjects (age range: 7.5–17.9 years, see Table 1 for details) were recruited in this study. The EOS patients were recruited from the Department of Psychiatry at the First Hospital of Shanxi Medical University, Taiyuan, China. The TDC participants were recruited from local communities in the same city. The Institutional Review Board at the First Hospital of Shanxi Medical University approved the study protocol. Written informed consent was obtained from each participant and the participant's guardian prior to data acquisition. The methods were carried out in accordance with the approved guidelines.
The EOS patients were diagnosed with a first episode of schizophrenia when recruited and they were drug-naïve. The diagnosis was made by at least two consultant psychiatrists according to the Diagnostic and Statistical Manual of Mental Disorders Fourth Edition (DSM-IV) criteria for schizophrenia51. Clinical symptoms of psychosis were quantified with PANSS52. The exclusion criteria were: 1) age >18 years, 2) history of neurological disease, brain injury, serious physical diseases, or other psychiatric disorders, 3) unsuitability for MRI scans (metal implants or claustrophobia), 4) history of taking any antipsychotic drugs, 5) history of substance abuse, 6) history of suicidal behavior, or 7) having a first-degree relative with a history of severe mental disorder or suicidal behavior. Two EOS patients and one TDC subject failed to complete the MRI scan and the data for four EOS patients and four TDC subjects were excluded by our quality control procedure (see below). Among the remaining patients, the PANSS scores for two patients were not available. Finally, 26 EOS patients and 25 TDCs are used for subsequent image analyses (See Table 1 for their demographical and clinical information).
Image acquisition, preprocessing and quality control
All imaging data were collected on a 3.0 T Siemens Trio MRI scanner at the First Hospital of Shanxi Medical University. Resting-state scans were acquired with an echo-planar imaging (EPI) sequence (32 axial slices, acquired from inferior to superior in an interleaved manner, FOV = 240 mm, matrix = 64 × 64, slice thickness = 4.0 mm, gap = 0.0 mm, TR/TE = 2500/30 ms, FA = 90°, 212 volumes, duration 8′50″) and anatomical scans were acquired with a T1-weighted 3D MP-RAGE sequence (160 continuous sagittal slices, slice thickness = 1.2 mm, FOV = 225 × 240 mm, matrix = 240 × 256, TR/TE/TI = 2300/2.95/900 ms, FA = 9°). Subjects were instructed to close their eyes and remain awake during the scan. After the scans, all subjects confirmed that they did not fall asleep during the scan.
The images were preprocessed using the Connectome Computation System (CCS: http://lfcd/psych.ac.cn/ccs.html)53. After motion and slice-timing correction, the resting-state data were band-pass filtered (0.01–0.1 Hz) and input to subject-level independent component analyses. Spatial transformations for normalization were estimated using boundary-based registration, combined with a nonlinear transformation between individual anatomical space and the MNI152 standard space using FNIRT in FSL. The transformations were then applied to the spatial maps derived from the independent components. For computation of functional connectivity, nuisance time series in white matter, ventricles and head motion parameters as represented in Friston-24 model54 were regressed out from voxel-wise time series. The residuals were spatially smoothed with a Gaussian kernel of 6 mm full-width-at-half-maximum before the band-pass temporal filtering.
A data quality control procedure was conducted to ensure the data were usable for subsequent analyses. The structural images were visually inspected for quality of tissue segmentation and brain registration. For functional images, in addition to the above procedures, subjects were excluded for excessive head motion, as defined by root-mean-square of frame-wise displacement55 larger than 0.2 mm.
gRAICAR analysis
The preprocessed functional images were processed using gRAICAR14,15,16 to characterize the consistency of the ICNs across all of the subjects. Details of the gRAICAR algorithm can be found in the original paper15 and Figure 1 in a more recent publication16 provides a more detailed illustration of gRAICAR algorithm. Briefly, the spatial independent components were derived using the MELODIC module of FSL56, where the number of independent components (ICs) was automatically determined. All of the ICs from the subjects of both groups were pooled in gRAICAR and normalized mutual information between every pair of ICs was computed to yield a full similarity matrix. The full similarity matrix was then searched to match ICs across different subjects, forming group-level ACs. Each AC was formed by a set of matched ICs containing one IC from each subject. For each of the ACs, a similarity matrix was computed to reflect the similarity between its comprising ICs, each representing a subject. In the inter-subject similarity matrix, the centrality of a subject's IC was computed by summing up the similarity metrics between that subject's IC and all other ICs in that AC. The centrality measures were then used as weights to average the spatial maps of the comprising ICs into a group-level spatial map for that AC. In summary, the spatial maps of the ACs represent ICNs or artifacts in the resting-state data and the similarity matrices reveal inter-subject consistency of the ACs.
Reliability of the gRAICAR findings
We employed a bootstrap approach to evaluate the likelihood that the ICN-symptom associations reported in this study can be found in a different sample of subjects, with the same sample size for patients and normal controls. The subjects were randomly sampled with replacement while keeping the original sample sizes for patients (n = 26) and healthy controls (n = 25) unchanged. In other words, for each bootstrap sample, 26 patients were randomly chosen from the original sample, allowing the same patient to be included multiple times. Similarly, 25 healthy controls were randomly selected from the original sample, allowing for replications of the selected subjects. This bootstrap sample thus contained the same numbers of patients and healthy controls as in the original dataset, but represented a different inter-subject variability.
gRAICAR was applied to this bootstrap sample to obtain ACs and their corresponding ICN-derived similarity matrices. The ACs corresponding to the original ICNs of interest (the PCU-AG ICN or the STG-IFG ICN described above) were selected by choosing the ones exhibiting the maximal spatial correlation coefficients (within all ACs obtained in the same bootstrap sample) with the original ICN maps reported in Figures 2 and 3. Meanwhile, the clinical similarity matrices were constructed based on the resampled subjects. The ICN-derived similarity matrices from the selected AC and the clinical similarity matrices were both transformed into vectors before their correlation coefficients, r, were computed. This procedure was repeated 5000 times to obtain the sample distributions of r. The mean values of the sample distributions approximate the population-level estimates of the associations between ICN- and symptom-derived inter-subject similarity matrices.
Subject community detection based on brain networks
A graphical demonstration of the subject community detection procedures is shown in Figure 6. The similarity matrix of each AC represents a weighted graph with subjects as nodes and the similarity between the spatial maps of their ICs' as weights of the edges. Each AC's graph was further investigated to detect subject communities based on the similarity of their ICs. The “k-clique” method was employed to conduct the community detection analyses in a data-driven manner so that the subject community was not affected by a priori groups57. In graph theory, a clique is defined as a sub-graph in which every two nodes are connected58. A maximal-clique is a clique that contains the maximum number of mutually connected nodes and cannot be extended by including more nodes (i.e., it's not part of a larger clique). The “k-clique” method identifies multiple overlapping maximal-cliques in a network and labels them as a community if their number of overlapping nodes is greater than a threshold, k.
Flowchart of community detection algorithm.
The procedures are demonstrated using eight subjects. For each AC obtained in gRAICAR, its similarity matrix (A) is represented using an undirected weighted graph (B), where the red nodes stand for the eight subjects and the distance between nodes indicates the similarities of the spatial maps from different subjects (shorter distance means higher similarity). The graph is then converted into a binary graph (C), where every two nodes are either connected or disconnected, by thresholding the edges with a probability threshold, p, derived from gRAICAR permutation tests. Within this binary graph, an algorithm is performed to search for maximal-cliques. In a maximal-clique, every pair of nodes is connected, indicating a dense cluster formed by the element nodes. In (D), three colors are used to mark three different maximal-cliques. The maximal-cliques are then merged if they share k nodes. A subject community is defined by the merged maximal-cliques (E).
Choosing the optimal thresholds
As demonstrated in Figure 6, the two parameters applied in the community detection model are the threshold for significant connections (p) and the threshold on the number of overlapping nodes for combining maximal-cliques into communities (k).
The first threshold, p, was used to convert the weighted subject graphs into binary graphs that represent whether a connection with significant strength exists between two subjects. To setup a proper value for this threshold, we first generated an empirical null-distribution based on the null hypothesis that ICs randomly selected to form a null-AC do not represent the same ICN. Specifically, we assigned a random IC from each subject to a null-AC and computed the similarity between subjects represented in the similarity matrix of the null-AC. This procedure was repeated 1000 times to generate a null-distribution of similarity metrics. The value at a certain percentile of the null distribution can be used as a statistical threshold (e.g., the value at the 99.5 percentile corresponds to p = 0.005) to eliminate any sub-threshold connections and generate the binary graph. To account for multiple comparison errors, we chose p = 0.01, 0.0075, 0.005 and 0.0025 as candidate values because when applying a smaller threshold of p = 0.001, more than 90% of the edges in the graphs disappeared, thus disabling further community detection. For the second parameter, k, the study originally proposing the method found k = 5 and 6 to be optimal57, since when k was greater than 6, the cliques were rarely merged. We therefore set the range of k from 2 to 6.
We used a reproducibility criterion to select the optimal combination of the two thresholds. The procedures are demonstrated in Figure 7. The output of the model is a profile of membership labeling subjects as 1 if they belong to a community and as 0 if they do not. We used cosine similarity to measure the similarity of the label profiles obtained under different choices of parameters. We first determined the optimal values of p by comparing the sum of all cosine similarity metrics among the label profiles obtained with different k values, under each choice of p value (k changed from 2 to 6, yielding 5 label profiles and 10 similarity metrics between every two profiles). After determining the optimal p value that produced the largest sum of cosine similarity, the optimal k value was determined by choosing the k value whose membership profile had the maximum sum of cosine similarities (i.e., most similar) to all other profiles from different values of k (under the optimal p value). Using the above criterion, we found that for both ICNs, an optimal combination of p = 0.005 and k = 5 produced the greatest sum of similarities across the subject community profiles. We also evaluated the influence of different parameters in the k-clique community detection algorithms. The methods and results are presented in supplementary materials.
Graphical demonstration of procedures to evaluate robustness to thresholds.
Two thresholds are used in the community detection algorithm: the probabilistic threshold, p, for binarizing the subject graph and the number of shared (overlapping) nodes between maximal-cliques, k, for merging the maximal-cliques into a subject community. For a given p, k values ranging from 2 to 6 are examined. For each choice of k, the community detection procedures are performed to generate a profile of subject membership, shown by the vectors indicating whether each subject is included in the subject community. Black color means a subject is in the community, while white color means a subject is not included. The cosine similarity between the profile vectors from different choices of k are computed to generated a similarity matrix. The sum along a given row indicates the reproducibility of that subject profile for a corresponding choice of k and the sum of the entire similarity matrix indicates the reproducibility of that subject profile for the given p. The p value with the maximal reproducibility value (sum of entire matrix) is first chosen and the optimal k is selected based on the maximal subject profile reproducibility (sum of row) under the given p.
References
Vyas, N. S., Hadjulis, M., Vourdas, A., Byrne, P. & Frangou, S. The Maudsley early onset schizophrenia study. Predictors of psychosocial outcome at 4-year follow-up. Eur. Child Adoles. Psy. 16, 465–470 (2007).
Rabinowitz, J., Levine, S. Z. & Hafner, H. A population based elaboration of the role of age of onset on the course of schizophrenia. Schizophr. Res. 88, 96–101 (2006).
Frazier, J. A. et al. Pubertal development and onset of psychosis in childhood onset schizophrenia. Psychiat. Res. 70, 1–7 (1997).
Hollis, C. Adult outcomes of child- and adolescent-onset schizophrenia: diagnostic stability and predictive validity. Am. J. Psychiat. 157, 1652–1659 (2000).
Vyas, N. S., Kumra, S. & Puri, B. K. What insights can we gain from studying early-onset schizophrenia? The neurodevelopmental pathway and beyond. Expert Rev. Neurother. 10, 1243–1247 (2010).
Frazier, J. A. et al. Brain anatomic magnetic resonance imaging in childhood-onset schizophrenia. Arch. Gen. Psychiat. 53, 617–624 (1996).
Rapoport, J. L. & Gogtay, N. Brain neuroplasticity in healthy, hyperactive and psychotic children: insights from neuroimaging. Neuropsychopharmacol. 33, 181–197 (2008).
Thompson, P. M. et al. Mapping adolescent brain change reveals dynamic wave of accelerated gray matter loss in very early-onset schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 98, 11650–11655 (2001).
Greicius, M. Resting-state functional connectivity in neuropsychiatric disorders. Curr. Opin. Neurol. 21, 424–430 (2008).
Lynall, M. E. et al. Functional connectivity and brain networks in schizophrenia. J. Neurosci. 30, 9477–9487 (2010).
Schmitt, A., Hasan, A., Gruber, O. & Falkai, P. Schizophrenia as a disorder of disconnectivity. Eur. Arch. Psy. Clin. N. 261, 150–154 (2011).
Kapur, S., Phillips, A. G. & Insel, T. R. Why has it taken so long for biological psychiatry to develop clinical tests and what to do about it? Mol. Psychiatr. 17, 1174–1179 (2012).
Insel, T. et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am. J. Psychiat. 167, 748–751 (2010).
Yang, Z., LaConte, S., Weng, X. & Hu, X. Ranking and averaging independent component analysis by reproducibility (RAICAR). Hum. Brain Mapp. 29, 711–725 (2008).
Yang, Z. et al. Generalized RAICAR: discover homogeneous subject (sub)groups by reproducibility of their intrinsic connectivity networks. Neuroimage 63, 403–414 (2012).
Yang, Z. et al. Connectivity trajectory across lifespan differentiates the precuneus from the default network. Neuroimage 89, 45–56 (2014).
Zapala, M. A. & Schork, N. J. Statistical properties of multivariate distance matrix regression for high-dimensional data analysis. Front. Genet. 3, 190 (2012).
Seiferth, N. Y. et al. Neuronal correlates of facial emotion discrimination in early onset schizophrenia. Neuropsychopharmacol. 34, 477–487 (2009).
Rapoport, J. L. et al. Progressive cortical change during adolescence in childhood-onset schizophrenia. A longitudinal magnetic resonance imaging study. Arch. Gen. Psychiat. 56, 649–654 (1999).
Jacobsen, L. K. et al. Quantitative morphology of the cerebellum and fourth ventricle in childhood-onset schizophrenia. Am. J. Psychiat. 154, 1663–1669 (1997).
Marquardt, R. K. et al. Abnormal development of the anterior cingulate in childhood-onset schizophrenia: A preliminary quantitative MRI study. Psychiat. Res-Neuroim. 138, 221–233 (2005).
Hagmann, P. et al. Mapping the structural core of human cerebral cortex. PLoS Biol. 6, e159 (2008).
van den Heuvel, M. P. & Sporns, O. Rich-club organization of the human connectome. J. Neurosci. 31, 15775–15786 (2011).
Sporns, O. The human connectome: a complex network. Ann. N. Y. Acad. Sci. 1224, 109–125 (2011).
Greicius, M. D., Krasnow, B., Reiss, A. L. & Menon, V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc. Natl. Acad. Sci. U. S. A. 100, 253–258 (2003).
Raichle, M. E. et al. A default mode of brain function. Proc. Natl. Acad. Sci. U. S. A. 98, 676–682 (2001).
Cavanna, A. E. The precuneus and consciousness. CNS Spectr. 12, 545–552 (2007).
Wallentin, M., Weed, E., Ostergaard, L., Mouridsen, K. & Roepstorff, A. Accessing the mental space-Spatial working memory processes for language and vision overlap in precuneus. Hum. Brain Mapp. 29, 524–532 (2008).
Buckner, R. L., Andrews-Hanna, J. R. & Schacter, D. L. The brain's default network: anatomy, function and relevance to disease. Ann. N. Y. Acad. Sci. 1124, 1–38 (2008).
Whitfield-Gabrieli, S. et al. Hyperactivity and hyperconnectivity of the default network in schizophrenia and in first-degree relatives of persons with schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 106, 1279–1284 (2009).
Kuhn, S. & Gallinat, J. Resting-state brain activity in schizophrenia and major depression: a quantitative meta-analysis. Schizophr. Bull. 39, 358–365 (2013).
Mitchell, R. L., Elliott, R. & Woodruff, P. W. fMRI and cognitive dysfunction in schizophrenia. Trends Cogn. Sci. 5, 71–81 (2001).
Puri, B. K. Progressive structural brain changes in schizophrenia. Expert Rev. Neurother. 10, 33–42 (2010).
Flaum, M. et al. Symptom dimensions and brain morphology in schizophrenia and related psychotic disorders. J. Psychiatr. Res. 29, 261–276 (1995).
Antonova, E., Sharma, T., Morris, R. & Kumari, V. The relationship between brain structure and neurocognition in schizophrenia: a selective review. Schizophr. Res. 70, 117–145 (2004).
Young, A. H. et al. A magnetic resonance imaging study of schizophrenia: brain structure and clinical symptoms. Br. J. Psychiat. 158, 158–164 (1991).
Roberts, G. W. Schizophrenia: a neuropathological perspective. Br. J. Psychiat. 158, 8–17 (1991).
Woodruff, P. W. et al. Structural brain abnormalities in male schizophrenics reflect fronto-temporal dissociation. Psychol. Med. 27, 1257–1266 (1997).
Giedd, J. N. et al. Childhood-onset schizophrenia: progressive brain changes during adolescence. Biol. Psychiat. 46, 892–898 (1999).
Jacobson, S. et al. Structural and functional brain correlates of subclinical psychotic symptoms in 11–13 year old schoolchildren. Neuroimage 49, 1875–1885 (2010).
Jacobson McEwen, S. et al. Resting-state connectivity deficits associated with impaired inhibitory control in non-treatment-seeking adolescents with psychotic symptoms. Acta Psychiatr. Scand. 129, 134–142 (2014).
Yung, A. R. & McGorry, P. D. The prodromal phase of first-episode psychosis: past and current conceptualizations. Schizophr Bull. 22, 353–370 (1996).
Yung, A. R. et al. Monitoring and care of young people at incipient risk of psychosis. Schizophr Bull. 22, 283–303 (1996).
Ziauddeen, H., Dibben, C., Kipps, C., Hodges, J. R. & McKenna, P. J. Negative schizophrenic symptoms and the frontal lobe syndrome: one and the same? Eur. Arch. Psy. Clin. N. 261, 59–67 (2011).
Peralta, V. & Cuesta, M. J. Psychometric properties of the positive and negative syndrome scale (PANSS) in schizophrenia. Psychiat. Res. 53, 31–40 (1994).
Lancon, C., Auquier, P., Nayt, G. & Reine, G. Stability of the five-factor structure of the Positive and Negative Syndrome Scale (PANSS). Schizophr. Res. 42, 231–239 (2000).
Borsook, D., Becerra, L. & Fava, M. Use of functional imaging across clinical phases in CNS drug development. Transl. Psychiatry. 3, e282 (2013).
Emsley, R., Rabinowitz, J., Torreman, M. & RIS-INT-35 Early Psychosis Global Working Group. The factor structure for the Positive and Negative Syndrome Scale (PANSS) in recent-onset psychosis. Schizophr. Res. 61, 47–57 (2003).
Andrews, G. et al. Dimensionality and the category of major depressive episode. Int. Method. Psych. 16 Suppl 1, S41–51 (2007).
Miller, J. What is the probability of replicating a statistically significant effect? Psychon. B. Rev. 16, 617–640 (2009).
Amercian Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-IV-TR. (American Psychiatric Publishing, Inc., 2000).
Kay, S. R., Fiszbein, A. & Opler, L. A. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr. Bull. 13, 261–276 (1987).
Zuo, X. N. et al. Toward reliable characterization of functional homogeneity in the human brain: preprocessing, scan duration, imaging resolution and computational space. Neuroimage 65, 374–386 (2013).
Friston, K. J., Williams, S., Howard, R., Frackowiak, R. S. & Turner, R. Movement-related effects in fMRI time-series. Magn. Reson. Med. 35, 346–355 (1996).
Patriat, R. et al. The effect of resting condition on resting-state fMRI reliability and consistency: a comparison between resting with eyes open, closed and fixated. Neuroimage 78, 463–473 (2013).
Beckmann, C. F. & Smith, S. M. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans. Med. Imaging 23, 137–152 (2004).
Palla, G., Derenyi, I., Farkas, I. & Vicsek, T. Uncovering the overlapping community structure of complex networks in nature and society. Nature 435, 814–818 (2005).
Luce, R. D. & Perry, A. D. A method of matrix analysis of group structure. Psychometrika 14, 95–116 (1949).
Acknowledgements
This study is supported by the National Key Technologies R&D Program of China (2012BAI36B01), the Natural Science Foundation of China (81270023, 81278412, 81171409, 81000583, 81220108014), the Foundation of Beijing Key Laboratory of Mental Disorders (2014JSJB03), the Intramural Research Program of the National Institute of Mental Health, the Key Research Program and the Hundred Talents Program of the Chinese Academy of Sciences (KSZD-EW-TZ-002) and the Program for New Century Excellent Talents in University. All of the authors declare no conflict of interest. The computations were performed on the NIH Biowulf Cluster System (http://biowulf.nih.gov) and the Dell Blade Cluster System at the Institute of Psychology, Chinese Academy of Sciences. We acknowledge the comments on the manuscript from Dr. F. Xavier Castellanos.
Author information
Authors and Affiliations
Contributions
Z.Y., Y.X. and X.Z. designed the study. Z.Y., T.X., G.C. and X.Z. analyzed the data. Z.Y., C.H., D.H., G.C., X.Z., G.N. and P.B. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Materials
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Yang, Z., Xu, Y., Xu, T. et al. Brain Network Informed Subject Community Detection In Early-Onset Schizophrenia. Sci Rep 4, 5549 (2014). https://doi.org/10.1038/srep05549
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05549
This article is cited by
-
Precision behavioral phenotyping as a strategy for uncovering the biological correlates of psychopathology
Nature Mental Health (2023)
-
Disassociated and concurrent structural and functional abnormalities in the drug-naïve first-episode early onset schizophrenia
Brain Imaging and Behavior (2022)
-
DisConICA: a Software Package for Assessing Reproducibility of Brain Networks and their Discriminability across Disorders
Neuroinformatics (2020)
-
Abnormalities of regional homogeneity and its correlation with clinical symptoms in Naïve patients with first-episode schizophrenia
Brain Imaging and Behavior (2019)
-
Multi-frequency Dynamic Weighted Functional Connectivity Networks for Schizophrenia Diagnosis
Applied Magnetic Resonance (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.