Abstract
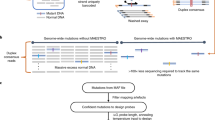
Fluorescent multiplex denaturing gradient gel electrophoresis (FMD) is a mutation screening technique designed to detect unknown as well as previously identified mutations. FMD constitutes a recent modification of the standard denaturing gradient gel electrophoresis (DGGE) technique, which combines multiplex PCR amplification of target DNA using fluorescently labeled primers with DGGE separation of the amplicon mixture, allowing immediate identification of sequence variants by wet gel scanning. FMD permits the simultaneous detection of small insertions, deletions and single nucleotide substitutions among multiple DNA fragments (up to 480 fragments) from 96 samples in parallel for each run. It increases output and reduces cost dramatically compared with classical DGGE, without sacrificing sensitivity and accuracy in detecting mutations. This protocol details an accurate, fast, nonradioactive and cost-effective way to screen the BRCA1 gene for mutations with high sensitivity, providing easily interpreted results. It may also be adapted to screen other target genes and/or used in large-scale epidemiological studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Meyer, J.M. & Ginsburg, G.S. The path to personalized medicine. Curr. Opin. Chem. Biol. 6, 434–438 (2002).
Amundadottir, L.T. et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 38, 652–658 (2006).
Jonson, J.A. & Evans, W.E. Molecular diagnostics as a predictive tool: genetics of drug efficacy and toxicity. Trends Mol. Med. 8, 300–305 (2002).
Sanger, F., Nicklen, S. & Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5467 (1977).
Sheffield, V.C., Beck, J.S., Kwitek, A.E., Sandstrom, D.W. & Stone, E.M. The sensitivity of single-strand conformation polymorphism analysis for the detection of single base substitutions. Genomics 16, 325–332 (1993).
Nataraj, A.J., Olivos-Glander, I., Kusukawa, N. & Highsmith, W.E. Jr. Single-strand conformation polymorphism and heteroduplex analysis for gel-based mutation detection. Electrophoresis 20, 1177–1185 (1999).
Hamzehloei, T., West, S.P., Chapman, P.D., Burn, J. & Curtis, A. Four novel germ-line mutations in the APC gene detected by heteroduplex analysis. Hum. Mol. Genet. 3, 1023–1024 (1994).
Zhang, Q. & Minoda, K. Mutation detection and genetic counseling in retinoblastoma using heteroduplex analysis. Jpn. J. Ophthalmol. 39, 432–437 (1995).
Fischer, S.G. & Lerman, L.S. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc. Natl. Acad. Sci. USA 80, 1579–1583 (1983).
Sheffield, V.C., Cox, D.R., Lerman, L.S. & Myers, R.M. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl. Acad. Sci. USA 86, 232–236 (1989).
Van Orsouw, N.J. & Vijg, J. Design and application of 2-D DGGE-based gene mutational scanning tests. Genet. Anal. 14, 205–213 (1999).
Wartell, R.M., Hosseini, S.H. & Moran, C.P. Jr. Detecting base pair substitutions in DNA fragments by temperature-gradient gel electrophoresis. Nucleic Acids Res. 18, 2699–2705 (1990).
Sorlie, T. et al. Mutation screening of the TP53 gene by temporal temperature gradient gel electrophoresis. Methods Mol. Biol. 291, 207–216 (2005).
Holinski-Feder, E. et al. DHPLC mutation analysis of the hereditary nonpolyposis colon cancer (HNPCC) genes hMLH1 and hMSH2. J. Biochem. Biophys. Methods 47, 21–32 (2001).
Pazaitou-Panayiotou, K. et al. Efficient testing of the RET gene by DHPLC analysis for MEN 2 syndrome in a cohort of patients. Anticancer Res. 25, 2091–2095 (2005).
Dolinsky, L.C.B., de Moura-Neto, R.S. & Falcao-Conceicao, D.N. DGGE analysis as a tool to identify point mutations, de novo mutations and carriers of the dystrophin gene. Neuromuscul. Disord. 12, 845–848 (2002).
van der Hout, A.H. et al. A DGGE system for comprehensive mutation screening of BRCA1 and BRCA2: application in a Dutch cancer clinic setting. Hum. Mutat. 27, 654–666 (2006).
Guldberg, P., Henriksen, K.F. & Guttler, F. Molecular analysis of phenylketonuria in Denmark: 99% of the mutations detected by denaturing gradient gel electrophoresis. Genomics 17, 141–146 (1993).
van Orsouw, N.J. et al. A highly accurate, low cost test for BRCA1 mutations. J. Med. Genet. 36, 747–753 (1999).
Bounpheng, M. et al. Rapid, inexpensive scanning for all possible BRCA1 and BRCA2 gene sequence variants in a single assay: implications for genetic testing. J. Med. Genet. 40, e33 (2003).
Schouten, J.P. et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 30, e57 (2002).
Hogervorst, F.B.L. et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 63, 1449–1453 (2003).
Miki, Y. et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266, 66–71 (1994).
Kuperstein, G., Jack, E. & Narod, S.A. A fluorescent multiplex-DGGE screening test for mutations in the BRCA1 gene. Genet. Test. 10, 1–7 (2006).
Bateman, J.F. et al. Reliable and sensitive detection of premature termination mutations using a protein truncation test designed to overcome problems of nonsense-mediated mRNA instability. Hum. Mutat. 13, 311–317 (1999).
De Benedetti, V.M. et al. Screening for mutations in exon 11 of the BRCA1 gene in 70 Italian breast and ovarian cancer patients by protein truncation test. Oncogene 13, 1353–1357 (1996).
Acknowledgements
The information for the primer sequences was kindly provided by Dr. Jan Vijg.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Zhang, S., Kuperstein, G. & Narod, S. Mutation screening using fluorescence multiplex denaturing gradient gel electrophoresis (FMD): detecting mutations in the BRCA1 gene. Nat Protoc 1, 3101–3110 (2006). https://doi.org/10.1038/nprot.2006.445
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.445
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.