Abstract
Activating transcription factor 4 (ATF4) is a critical mediator of metabolic and oxidative homeostasis and cell survival. ATF4 is elevated in response to diverse microenvironmental stresses, including starvation, ER stress damages and exposure to toxic factors. Here we show that ATF4 expression fosters the malignancy of primary brain tumors (WHO grade III and IV gliomas) and increases proliferation and tumor angiogenesis. Hence, ATF4 expression promotes cell migration and anchorage-independent cell growth, whereas siRNA-mediated knockdown of ATF4 attenuates these features of malignancy in human gliomas. Further experiments revealed that ATF4-dependent tumor promoting effects are mediated by transcriptional targeting the glutamate antiporter xCT/SCL7A11 (also known as system Xc-). Thus, xCT is elevated as a consequence of ATF4 activation. We further found evidence that ATF4-induced proliferation can be attenuated by pharmacological or genetic xCT inhibition and ferroptosis inducers such as sorafenib, erastin and GPx4 inhibitor RSL3. Further, fostered xCT expression promotes cell survival and growth in ATF4 knockdown cells. Moreover, increased xCT levels ameliorate sorafenib and erastin-induced ferroptosis. Conversely, ATF4 knockdown renders cells susceptible for erastin, sorafenib and RSL3-induced ferroptosis. We further identified that ATF4 promotes tumor-mediated neuronal cell death which can be alleviated by xCT inhibition. Moreover, elevated ATF4 expression in gliomas promotes tumor angiogenesis. Noteworthy, ATF4-induced angiogenesis could be diminished by ferroptosis inducers erastin and by GPx4 inhibitor RSL3. Our data provide proof-of-principle evidence that ATF4 fosters proliferation and induces a toxic microenvironmental niche. Furthermore, ATF4 increases tumor angiogenesis and shapes the vascular architecture in a xCT-dependent manner. Thus, inhibition of ATF4 is a valid target for diminishing tumor growth and vasculature via sensitizing tumor cells for ferroptosis.
Similar content being viewed by others
Introduction
Primary brain tumors are characterized by their specific microenvironmental niche containing pathological vessels and extracellular factors. These features promote angiogenesis and neuronal damage.1, 2 These stress conditions can, in principle, also affect tumor cells. However, tumor cells are able to withstand these factors and remain metabolically adapted under such toxic conditions. The homeostatic adaptation is mediated via the so-called integrated stress response (ISR).3 The ISR is critical for tumor survival and mediates its effects via protein synthesis reduction and enhanced activating transcription factor 4 (ATF4) expression.4, 5 Activating transcription factors belong to the family of basic region leucine zipper proteins and operate as key transcriptional regulators. ATF4 in particular is a member of the CREB/ATF family and ubiquitously expressed in human organs and can be activated in response to various stress signals such as anoxia, hypoxia, endoplasmic reticulum stress, amino acid deprivation and oxidative stress.6, 7 Stress-dependent expression of ATF4 causes adaptive responses in cells through regulating the expression of target genes. Noteworthy, ATF4-driven genes are mainly involved in amino acid synthesis, differentiation, metastasis, angiogenesis and drug resistance.8, 9, 10 A general phenomenon is that elevated ATF4 expression is often observed in human tumor pathologies.11, 12 Induction of the ISR requires ATF4 expression, and in this way tumor cells are believed to adapt and fit to their own created neurotoxic microenvironment.7, 13 Highly elevated ATF4 expression is hypothesized to facilitate tumor progression. In fact, transcription of many essential genes involved in tumor cell proliferation is regulated by ATF4.11, 14, 15 However, there are various effector genes induced by ATF4,16, 17 whereas it is still not elucidated which of them are operational and necessary for the malignancy and tumor progression.
In this study, we found that human cells show increased signs of malignancy induced by concomitant ATF4 expression. Conversely, ATF4 inhibition can attenuate tumor growth. Further, we found that ATF4 upregulates xCT and elevated xCT expression can rescue the ATF4 deletion phenotype. Conversely, ATF4-induced tumor growth can be reverted by inhibition of xCT. On the basis of these findings, we further investigated the role of ATF4 in the brain microenvironment. Here we found that ATF4 promotes tumor-induced cell death, which can be attenuated by xCT inhibition. Moreover, fostered ATF4 expression increases tumor angiogenesis and vessel growth. Our results indicate that ATF4 acts as a promoter of cellular resistance towards ferroptosis and cytotoxic stress, thereby promoting tumor malignancy.
Results
ATF4 fosters tumor cell growth
First, we investigated the expression levels of ATF4 in a variety of human tumors (NIH Rembrandt Glioma database). We found that ATF4 is significantly elevated in malignant gliomas compared to non-transformed human brain tissue (Figure 1a). Patients with high ATF4 expression levels displayed reduced overall survival time, while patients with low ATF4-expressing gliomas showed prolonged survival. (Figure 1b). We also found that the levels of ATF4 mRNA were significantly upregulated in malignant glioma samples (Figure 1c). Thus, gliomas with World Health Organization (WHO) classification grade II appeared with a two-fold ATF4 mRNA increase compared to normal brain samples, whereas most malignant gliomas (gliomas WHO° IV) exhibited a five-fold increase in ATF4 mRNA expression in comparison to normal brain tissue, respectively (Figure 1c). Given the fact that the ATF4 gene is often genetically amplified and overexpressed in human malignant gliomas, we next investigated whether alteration of the transcriptional master regulator ATF4 affects cell morphology and cell death. First, we investigated the endogenous ATF4 mRNA levels in various glioma cells such as T98G, U87 and U251 (Supplementary Figure 1a). There we found that U87 cells show lower ATF4 expression compared to U251 and T98 wild-type glioma cells, whereas U251 showed highest ATF4 mRNA levels (Supplementary Figure 1a). This prompted us to continue our investigation with U87 and U251 cells. To gain insights into this, we expressed and repressed ATF4 in human cells (Figures 1d–f). ATF4 was stably transfected into U87 and U251 glioma cells (ATF4OE), and ATF4 was depleted in human cells by RNA interference (ATF4KD). ATF4 mRNA levels were detected by quantitative RT-PCR (Figure 1e–f). Further cytoskeletal immuno-analysis of human glioma cells revealed that ATF4 has a strong impact on the cell morphology (Figure 1d). ATF4-overexpressing (ATF4OE) cells display a polymorphic shape and their cell bodies are significantly bigger than in ATF4KD and control cells (Figure 1d). In contrast, ATF4KD cells appear generally smaller and show a spindle-like phenotype with maximal two membrane extensions (Figure 1d).
ATF4 expression is associated with human malignant gliomas and changes the morphology of glioma cells. (a) Oncomine database analysis of different human primary brain tumor entities. Expression of ATF4 mRNA is higher in astrocytoma (n=148), GBM (n=228) and oligodendroglioma (n=67) compared to that of non-tumor tissue (n=28). ***P<0.001, Student’s t-test. (b) Kaplan–Meier survival analysis (Rembrandt Glioma Dataset) of high versus low ATF4-expressing gliomas. P<0.001, log-rank test. (c) ATF4 mRNA and xCT mRNA in human glioblastoma (WHO° II, n=12; WHO° IV, n=15) and normal brain (con, n=3) samples were evaluated by RT-PCR. ***P<0.001, Student’s t-test. (d) Morphology of U87 cells expressing GFP (ctrl), pSuper-GFP (sh-ctrl), ATF4 overexpression (ATF4OE) and shRNA-mediated ATF4 knockdown (ATF4KD) stained for actin (red) and nuclei (Hoechst 33342, blue). Scale bar represents 50 μm. ATF4OE cells show polymorphic cell shape and are generally bigger in size than ATF4KD and control cells. ATF4KD glioma cells are smaller compared to ATF4OE and control cells. (e, f) Quantification of ATF4 mRNA expression in ATF4 overexpression and shRNA-mediated knockdown glioma cells (U87, U251) via quantitative RT-PCR (n=3). Statistical significance was calculated with Student’s t-test (mean±s.d., ***P<0.001).
We further analyzed the cell proliferation and cell viability in ATF4 challenged cells. ATF4OE glioma cells significantly increased cell growth compared to control cells (ctrl), whereas ATF4 rknockdown resulted in 40% growth reduction (Figures 2a and b). Hence, we investigated the impact of ATF4 expression on the cell cycle using BrdU and 7-AAD double staining. We compared the cell cycle profiles of controls, ATF4OE and ATF4KD cells, and found a significantly prolonged G2/M phase in ATF4KD cells, whereas ATF4OE cells showed ~10% increase in the S phase (Figure 2c). Thus, a reduction in the proliferation rate of ATF4KD cells could be caused due to an increased number of cells arrested in the G2/M phase.
ATF4 promotes glioma cell growth. (a) ATF4 depletion slowed down glioma cell growth. Cell growth was determined by MTT assay (n=8). Statistical significance was calculated with Student’s t-test (mean±s.d., **P<0.01; ***P<0.001). (b) Quantification of cell growth over time. 5000 cells were seeded per well in 12-well plates and counted for 3 days (n=6). Statistical significance was calculated with Student’s t-test (mean±s.d., **P<0.01; ***P<0.001). (c) Cell cycle profile of ATF4OE and ATF4KD U87 cells was monitored with 7-AAD (n=3) (bottom). Quantification of the sub G1, G1, S and G2/M percentage in ATF4OE, ATF4KD, and respective control cells shown in (c). Statistical significance was calculated with Student’s t-test (given are mean±s.d., *P<0.05; **P<0.01; ***P<0.001). BrdU incorporation and 7-AAD staining of ATF4OE and ATF4KD U87 cells was analyzed by flow cytometry (n=3). ATF4KD cells showed prolonged G2/M phase.
Next, we investigated the anchorage-independent cell growth as a characteristic of oncogenic transformation and sign of malignancy. Quantification of colony formation in soft agar revealed that ATF4 overexpression resulted in a nearly 1.5-fold increase of colonies (Figures 3a and b). In contrast, ATF4 suppression led to a two-fold reduction in colony numbers compared to control cells. (Figures 3a and b). We continued characterizing the functional impact of ATF4 by studying the migration activity. For that, we cultured cells in a 3D spheroid matrix and monitored the invasive growth front (Figures 3c and d). We observed that ATF4OE cells migrated significantly faster than control cells (Figures 3c and d). Conversely, ATF4KD cells showed a low invasive growth pattern and ATF4KD cells migrate significantly slower (Figures 3c and d). Altogether, these results demonstrate that the ATF4 expression promotes glioma cell proliferation and migration.
ATF4 fosters anchorage-independent cell growth and migration. (a) Soft agar colony-forming assay with controls (crtl), ATF4OE and ATF4KD U87 cells. Representative images show colonies after 21 days post seeding. Scale bar represents 100 μm. (b) Quantitative analysis of colony formation in soft agar. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=3, ***P<0.001). (c) Representative microscopic images of glioma spheroids on day 1, day 2, day 4 and day 6. (d) Quantification of spheroid area revealed that ATF4OE cells migrate faster than control cells, but significantly slower in ATF4KD cells. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=6, *P<0.05; **P<0.01; ***P<0.001).
ATF4 regulates xCT expression and glutamate secretion
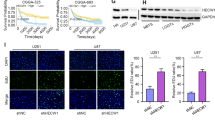
ATF4 is a potent inducer of ISR related and antioxidant gene expression. So far known transcriptional targets of ATF4 are xCT, HO-1, GPx1, SOD1 and others.18 We tested, whether ATF4 can drive the expression and function of the glutamate antiporter xCT (SCL7a11/SCX) since xCT is a key player in brain tumor-induced intoxication of the microenvironment.19, 20 We found that the levels of xCT mRNA were significantly increased in human malignant glioma specimens (Figure 4a). Gliomas WHO° II appeared with a five-fold xCT mRNA increase compared to normal brain samples, whereas gliomas WHO° IV exhibited a 19-fold increase in xCT mRNA expression in comparison to normal brain tissue, respectively (Figure 4a). We then found that fostered ATF4 expression upregulated significantly the glutamate transporter xCT mRNA (Figure 4b). Moreover, increased xCT protein levels were accompanied by enhanced levels of ATF4 (Figure 4c). In contrast, ATF4 knockdown in human cells markedly reduced the xCT expression (Figures 4b and c). To validate this finding, we analyzed the extracellular glutamate and cystine levels secreted by cultured cells. Noteworthy, ATF4OE cells secreted higher amounts of glutamate into the extracellular space (Figure 4d). In comparison, knockdown of ATF4 markedly reduced glutamate release and cystine uptake, presumably because of a decrease in intracellular glutamate available for transamination (Figure 4d). Given that xCT antiporter activity was determined by extracellular glutamate levels, we found elevated glutamate concentrations in ATF4OE cells (Figure 4d). In contrast, knockdown of ATF4 significantly reduced the xCT transporter activity (Figure 4d).
ATF4 promotes glioma cell proliferation and glutamate secretion through xCT regulation. (a) xCT mRNA in human glioblastoma (WHO° II, n=12; WHO° IV, n=15) and normal brain (con, n=3) samples was evaluated by RT-PCR. ***P<0.001, Student’s t-test. (b) RT-PCR analysis of xCT mRNA levels in ATF4OE and ATF4KD glioma cells. Statistical significance was calculated with Student’s t-test (given is mean±s.d., n=3, *P<0.05; **P<0.01; ***P<0.001). (c) Immunoblot analysis of xCT, ATF4 and GAPDH (served as a loading control) in U87 and U251 cells. (d) Stable transfected ATF4OE and ATF4KD U87 cells were cultured in glutamate-free medium for 12 h, after which the culture supernatants were assayed for glutamate and cystine by HPLC. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=3, ***P<0.001). (e) Glioma cells were co-transfected with ATF4-overexpressing plasmid plus xCT shRNA or ATF4 shRNA plus xCT-overexpressing constructs. Cell survival was determined by MTT assay. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=8, *P<0.05; **P<0.01; ***P<0.001). (f) ATF4OE cells were treated with xCT blocker S-4-carboxy-phenylglycine (S4CPG) and sulfasalazine (SAS). Cell survival was determined by MTT assay. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=8, *P<0.05; **P<0.01; ***P<0.001; n.s., no significance).
Next, we examined whether the ATF4 target gene xCT is required for cell growth and proliferation. For this, we knocked down xCT, in particular in ATF4OE and upregulated xCT expression in ATF4KD cells (Figure 4e). Silencing of xCT expression by RNAi severely limited ATF4OE cell growth below the levels of controls (Figure 4e). These findings could be confirmed in two independent human cell lines (Figure 4e). Interestingly, xCT expression in the background of ATF4KD cells could fully rescue the growth and proliferation rates and reinstalled the levels of controls (Figure 4e). We further confirmed these findings by targeting xCT with a pharmacological approach (Figure 4f). Interestingly, increased proliferation of ATF4OE cells could be inhibited by xCT-specific inhibitors S-4-carboxy-phenylglycine (S-4CPG) and the clinically established drug sulfasalazine (SAS) (Figure 4f).21 However, we found that xCT-specific inhibitors had no inhibitory growth effects on control cells, indicating that ATF4-driving cell growth is mediated by xCT (Figure 4f).
ATF4 expression induces acquired cell death resistance
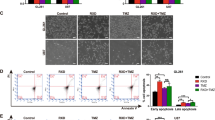
We further investigated the mechanisms of ATF4-driven proliferation by studying cytotoxic efficacy of chemotherapeutics. The rationale comes from the fact that chemotherapeutic agents such as sorafenib and erastin are also xCT inhibitors. xCT is a major player in iron-dependent non-apoptotic cell death.22 Proliferation assays and cell death monitoring revealed that 72 h after treatment sorafenib and erastin inhibited cell growth in a dose-dependent manner in controls (Figures 5a–f, Supplementary Figure 1b). Of note, ATF4OE cells were resistant to sorafenib and erastin compared to controls (Figures 5a–f). Conversely, ATF4KD cells became susceptible towards erastin and sorafenib-dependent cell growth inhibition (Figures 5a–f). Hence, we evaluated the effects of sorafenib and erastin by monitoring the annexin V/7-AAD apoptosis markers (Figure 5). Annexin V-positive/7-AAD-negative cells are defined as early apoptotic cells and annexin V-positive/7-AAD-positive cells are regarded as late apoptotic and early necrotic cells. The treatment of sorafenib and erastin in glioma cells significantly increased the pool of apoptotic cells in ATF4KD cells (Figures 5e and f). Meanwhile, ATF4KD cells displayed a moderate increase of necrotic cells compared to ATF4OE cells (Figures 5e and f) based on the annexin V-negative/7-AAD-positive characteristics defined as necrotic cell population. Similarly, the number of early apoptosis events was increased in ATF4KD cells compared to controls (Figures 5e and f). Altogether, these results demonstrate that ATF4 confers a multidrug-resistance phenotype in human cells and targeting ATF4 provides a method for increasing chemotherapeutic susceptibility in glioma cells.
ATF4 overexpression renders gliomas resistant to sorafenib and erastin. (a) ATF4OE, ATF4KD and Control U87 cells were treated with sorafenib wat a concentration of 2.5 μM, 5 μM and 10 μM for 3 days. PI staining (white dots) shows the amount of dead cells. Scale bar represents 200 μm. (b) Cell survival was measured by MTT in U87 and U251 cells. Sorafenib treatment resulted in a dose-dependent inhibition of cell viability. ATF4KD glioma cells were more sensitive to sorafenib. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=8, *P<0.05; **P<0.01; ***P<0.001). (c) ATF4OE, ATF4KD and control U87 cells were treated with 1, 2 and 5 μM erastin for 3 days. PI staining displys the amount of dead cells at the time of the MTT assay. Scale bar represents 200 μm. (d) Quantification of cell viability measured by MTT in U87 and U251 cells. ATF4OE glioma cells were resistant to erastin. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=8, *P<0.05; **P<0.01; ***P<0.001). (e, f) Treatment of ATF4OE, ATF4KD and control U87 cells with 10 μM sorafenib and 5 μM erastin for 72 h. Cell death was determined by flow cytometry using 7-AAD and Annexin V. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=3, *P<0.05; **P<0.01; ***P<0.001; n.s., no significance).
ATF4 knockdown renders cells susceptible for ferroptosis
In order to investigate whether ATF4 affects the induction of ferroptosis, we analyzed the cell death mechanisms induced by sorafenib or erastin with and without iron chelation (Figures 6 and 7, Supplementary Figure 2). U87 cells were exposed to sorafenib (10 μM) and erastin (10 μM) for 18 h. We found that sorafenib robustly induced cell death in ATF4KD U87 cells (Figures 6a–c). The sorafenib-induced cell death response could be almost completely rescued by the ferroptosis inhibitors deferoxamine (DFO; 100 μM) and ferrostatin-1 (Fer-1; 0.5 μM) (Figures 6d and e, Supplementary Figure 2). DFO and Fer-1 both significantly reversed the cell viability of ATF4KD U87 cells in response to sorafenib (Figure 6b, Supplementary Figure 2). In contrast, sorafenib-induced cell death in ATF4OE U87 cells could not be rescued by iron chelators (Figure 6d). A similar pharmacological response profile was found when the reference inducer of ferroptosis, erastin, was applied (Figures 7a–c). Flow cytometric analysis using the fluorescent probes C11-BODIPY demonstrated that ATF4KD U87 cells showed increased lipid peroxidation and increased sensitivity towards sorafenib and erastin (Figures 6c and 7c). In contrast, ATF4OE U87 cells did not show raised lipid peroxidation when treated with sorafenib and erastin (Figures 6c and 7c). We further studied the effects of the glutathione peroxidase 4 (GPx4) inhibitor and ferroptosis inducer RSL3.23 Consistent with these data, RSL3 treatment (at 0.1 μM) for 18 h significantly induced cell death and growth inhibitory effect in U87 cells, and this effect was strongest in ATF4KD cells (Figures 8a and b). Noteworthy, DFO and Fer-1 were both effective in preventing RSL3-induced ferroptosis in ATF4OE and ATF4KD U87 cells (Figures 8a and b). We also observed that RSL3-induced cell death was associated with the accumulation of lipid peroxidation. Treatment of ATF4KD U87 cells with RSL3 resulted in obvious increase in lipid peroxidation which was suppressed by co-treatment with DFO and Fer-1, whereas lipid peroxidation in ATF4OE U87 cells increased slightly (Figure 8c).
ATF4 knockdown renders cells susceptible for ferroptosis and lipid ROS accumulation. (a) Representative images of ATF4OE and ATF4KD U87 cells treated with 10 μM sorafenib (S) with or without deferoxamine (DFO, 100 μM) or ± ferrostatin-1 (Fer-1, 0.5 μM) for 18 h. Morphological changes were observed, including cell rounding and shrinkage. Scale bar represents 100 μm. (b) Cell survival was measured by MTT in U87 cells. Ferroptosis inhibitors rescued ATF4KD cell proliferation inhibited by sorafenib but not in ATF4OE cells. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=8, ***P<0.001; n.s., no significance). (c) Lipid ROS accumulation was assessed in U87 cells treated for 18 h with sorafenib ± DFO or ± Fer-1 using C11-BODIPY. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=3, *P<0.05; **P<0.01; ***P<0.001; n.s., no significance). (d) PI staining shows the amount of dead cells following treatment. Ferroptosis inhibitors prevented sorafenib-induced cell death. Scale bar represents 200 μm. (e) Effect of sorafenib ± DFO, ± Fer-1 on glioma (U87) cell death for 18 h as assessed by flow cytometry using 7-AAD and Annexin V. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=3, *P<0.05; **P<0.01; ***P<0.001; n.s., no significance).
Inhibition of ferroptosis rescue erastin-induced cell death in ATF4 knockdown cells. (a) Visualization of ATF4OE and ATF4KD U87 cells treated with 10 μM erastin (E) ± DFO (100 μM) or ± Fer-1 (0.5 μM) for 18 h. Scale bar represents 100 μm. (b) Quantification of cell viability measured by MTT in U87 cells. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=8, *P<0.05; **P<0.01; ***P<0.001; n.s., no significance). Ferroptosis inhibitors rescue erastin-induced cell death in ATF4KD Statistical significance was calculated with Student’s t-test (*P<0.05; n.s., no significance). (c) Lipid ROS production was assessed in U87 cells treated for 18 h with erastin ± DFO or ± Fer-1 by flow cytometry using C11-BODIPY. Statistical significance was calculated with Student’s t-test (n=3). (d) PI staining (white dots) reveals dead cells in erastin-treated ATF4OE and ATF4KD U87 cells. Scale bar represents 200 μm. (e) Effects of erastin ± DFO, ± Fer-1 on glioma (U87) cells assessed by flow cytometry using 7-AAD and Annexin V. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=3, *P<0.05; **P<0.01; ***P<0.001; n.s., no significance).
RSL3-induced accumulation of lipid ROS and ferroptosis in ATF4KD cells. (a) Representative images of ATF4OE and ATF4KD U87 cells treated with 0.1 μM RSL3 (R) ± DFO (100 μM) or ± Fer-1 (0.5 μM) for 18 h. Scale bar represents 100 μm. (b) Quantification of cell viability measured by MTT in U87 cells. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=8, *P<0.05; **P<0.01; ***P<0.001; n.s., no significance). Ferroptosis inhibitors counteracted the cell growth inhibitory effects of RSL3. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=8, ***P<0.001). (c) Lipid ROS production in U87 cells treated with RSL3 ± DFO or ± Ferr-1 for 8 h assayed by flow cytometry using C11-BODIPY. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=3, *P<0.05; **P<0.01; ***P<0.001; n.s., no significance).
Altogether, the increase in intracellular reactive oxygen species (ROS) affected glioma cell death and detachment (Figures 6a and d; Figures 7a and d; Figure 8a). In summary, these data show that sorafenib and erastin-induced cell death in ATF4 knockdown cells is associated with increased levels of ROS. ROS accumulation and subsequent cell death could be prevented by iron chelation of cellular iron uptake. Thus, our data evidence that ATF4 depletion renders cells susceptible for ferroptosis.
ATF4 overexpression increases neuronal damage through elevated xCT expression
To further investigate the functional relevance of ATF4 in glioma biology, we employed the vascular organotypic brain slice culture system termed VOGiM.24, 25 The VOGiM approach allows monitoring cell invasion and neuronal cell death in an organotypic microenvironment reflecting the in vivo situation in the brain. We found that ATF4-overexpressing cells impaired the neuronal integrity and promoted neuronal cell death compared to ATF4KD and control cells (Figures 9a and b). In addition, ATF4OE cells displayed increased tumor volume compared to controls (Figure 9c). These data confirm the in vitro findings in single cells reported in Figure 3.
ATF4 expression in gliomas mediates neuronal cell death. (a) Representative immunofluorescence images of organotypic brain slice culture system with implanted ATF4OE, ATF4KD and control U87 tumors. Neuronal cell death in living rain tissue was assessed by PI staining on day 5 in ex vivo. Scale bar represents 1 mm. (b) Quantification of tumor-induced neuronal cell death in surrounding healthy brain parenchyma. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=6, *P<0.05; **P<0.01; ***P<0.001; n.s., no significance). (c) Quantification of tumor growth in organotypic brain slice on the indicated day. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=4, *P<0.05; **P<0.01; ***P<0.001; n.s., no significance). (d) Primary rat neuronal cells were treated with supernatants from ATF4OE, ATF4KD, ATF4OE+xCTKD, ATF4KD+xCTOE and control cells (ctrl or sh-ctrl). Representative immunofluorescence images of β III-tubulin (green), GFAP (red) and Hoechst (blue) stained brain slices for detecting neurons, astrocytes and nuclei, respectively. PI staining shows the amount of dead cells after incubation with conditioned media for 24 h. Scale bar in upper panels represents 100 μm. Scale bar in lower panels represents 200 μm. (e) Quantification of cell death of primary rat neuronal cells after treatment with conditioned media from ATF4OE, ATF4KD and control cells. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=12, *P<0.05; **P<0.01; ***P<0.001; n.s., no significance).
Due to the identification of ATF4-mediated xCT expression in glioma cells, we aimed at validating the role of ATF4-xCT pathway in the context of tumor-induced neuronal damage. For this, we treated primary neurons for 24 h with conditioned supernatants from ATF4OE, ATF4KD, ATF4OE/xCTKD, ATF4KD/xCTOE and control cells. Cell death staining revealed that conditioned media from ATF4OE cells induced significantly more cell death in comparison to the effects from control supernatants (Figures 9d and e). To investigate whether such toxic effects are due to ATF4-mediated xCT expression, we tested conditioned media from ATF4OE/xCTKD cells. We found that conditioned media from ATF4OE/xCTKD cells was significantly weaker in inducing cell death compared to ATF4OE cells (Figures 9d and e). Similarly, neuronal cell death was increased when neurons were cultured in the presence of ATF4KD/xCTOE media compared in ATF4KD media (Figures 9d and e). These results show that xCT plays also a critical role in executing the ATF4-mediated neuronal cell death.
ATF4 promotes tumor angiogenesis
We further analyzed whether ATF4 has an impact on other hallmarks of tumor microenvironmental processes. Therefore, we investigated peritumoral vessel formation as an independent malignant feature of gliomas. We monitored peritumoral vessels, which were surrounding gliomas. We found that ATF4OE human glioma cells (U87 and U251) enhanced angiogenesis with increased tumor vessels and vessel length compared to control gliomas (Figures 10a and b). Interestingly, ATF4KD U251 cells showed reduced vessel nodes compared to controls (Figures 10a and b). These data indicate that ATF4 promotes tumor angiogenesis in gliomas.
ATF4 expression promotes tumor angiogenesis. (a) Representative immunofluorescence images displaying tumor vessels. Laminin immunostaining (red) was facilitated for detecting vessels surrounding gliomas after 5 days in culture. Upper panel shows representative images of GFP-positive U251 cells (green) and vessels (red) in the peritumoral area. Scale bar represents 200 μm. Lower panel shows higher magnification of vessels (red) within the peritumoural area. Scale bar represents 100 μm. (b) Quantification of vessels number and vessels length in the peritumoral area. ATF4OE U87 tumors (left histogram) and ATF4OE U251 tumors (right histogram) show increased vessel numbers and vessel lengths in the peritumoral area compared to control tumors. Conversely, ATF4KD tumors reveal reduced tumor vessels in the peritumoral area. Statistical significance was calculated with Student’s t-test (shown is mean±s.d., n=12, *P<0.05; **P<0.01; ***P<0.001; n.s., no significance).
Next, we sought to investigate the potential role of ferroptosis in ATF4-driven angiogenesis. For this, we performed the VOGiM approach with ATF4OE, ATF4KD and control glioma cells were treated with erastin (10 μM), RSL3 (5 μM) and Fer-1 (0.5 μM). Five days after treatment, erastin and RSL3 significantly inhibited peritumoral vessel formation surrounding ATF4KD and control gliomas (Figure 11). In contrast, in ATF4OE induced angiogenesis these angiogenesis reversing effects were lower but significant (Figures 11a and b). Fer-1, however, attenuated erastin and RSL3-suppressed angiogenesis of ATF4KD and control gliomas (Figures 11a and b). These data indicate that ATF4-induced angiogenesis can in part be reduced by erastin and RSL3-mediated ferroptosis.
ATF4-induced tumor angiogenesis can be diminished by inducing ferroptosis. (a) Control, ATF4OE, ATF4KD U87 cells (blue, white circle delineated) were implanted in rat brain slices and afterwards treated with erastin (10 μM) and RLS3 (5 μM) ± Fer-1 (0.5 μM) for 5 days. Erastin and RSL3 inhibited angiogenesis in control and ATF4KD gliomas and this effects could be reversed by Fer-1. Vessels were stained for laminin (red) and nuclei were stained by Hoechst (blue). Scale bar represents 100 μm. (b) Quantitative analysis of vessel numbers and vessels lengths in peritumoral area. The data are presented as mean±s.d. with n=3 and P-values set as: *P<0.05; **P<0.01; ***P<0.001; n.s. no significance.
Discussion
Here we present evidence that concomitant ATF4 expression promotes malignancy features of tumor cells such as cell growth, migration, angiogenesis and acquired chemo-resistance. One main downstream target could be identified as the glutamate antiporter xCT. Accumulating evidence has shown that xCT plays a critical role in developing and promoting malignancy of different cancer entities and in acquired chemo-resistance.11, 20, 26 Moreover, xCT has been discovered as a key player in ferroptosis, a special form of iron-dependent cell death with therapeutic implications.22, 27 Our results show that ATF4 conveys resistance to ferroptosis-mediated cell death. ATF4 is a critical mediator of metabolic and oxidative homeostasis. Also, ATF4 is implicated in cell survival and is elevated in response to ISR induced by diverse microenvironmental stresses, including starvation, ER stress damages and toxic exposure.28 Here we found that ATF4 is critically involved in human cell proliferation, invasive tumor growth and angiogenesis.
The novelty of this report is the identification of the glutamate antiporter xCT as a major driver of ATF4 functions. In fact, xCT expression has an important role in determining the malignancy in gliomas and other cancer entities.20, 29, 30, 31 Moreover, malignant gliomas induce neuronal degeneration and tumor-associated brain edema in a xCT-dependent manner.19 One major contributor to this glioma effects could be identified as the excitatory amino acid glutamate.20 The amino acid transporter system x with the catalytic domain xCT has been shown to be vital in releasing glutamate into the extracellular space. Since tumor-derived glutamate release is one major factor contributing to tumor-associated neurodegeneration and subsequent brain swelling, xCT is a prime target for therapeutic interventions in gliomas.32, 33 Thus, we addressed the question whether ATF4-induced alterations in cell growth and in the tumor microenvironment are causally linked to its transcriptional target xCT.19 Our analyses revealed that siRNA-mediated xCT silencing in ATF4OE cells can normalize cell proliferation. Moreover, deletion of ATF4 was sufficient in reducing proliferation and tumor growth. These effects could be abolished by reconstituting xCT in ATF4KD cells, demonstrating that xCT operates through glutamate release as an ATF4 executor. In this respect, it is no surprise that ATF4KD cells renders highly susceptible towards ferroptotic cell death. Our data reveal an important crosstalk of ATF4 and its downstream target xCT in gliomas. ATF4 confers ferroptosis resistance by its transcriptional regulation of xCT.
Finally, our data demonstrate that targeting ATF4 inhibits key features of gliomas to form a deadly microenvironment. First, ATF4 expression in gliomas fosters tumor-induced cell death of adjacent normal brain cells. This is an important hallmark of gliomas, which determines its malignancy.19, 20 Second, we found that ATF4 expression in gliomas increases tumor angiogenesis. Previous reports indeed showed that ATF4 is involved in tumor angiogenesis.34, 35 Up to now, the angiogenesis-promoting effects of ATF4 were linked to increased HIF1a and VEGF expression.36, 37 We could identify that xCT is a transcriptional target of ATF4 in glioma cells, and ATF4 promotes glioma cell growth through the functional activity of xCT, that is, increased glutamate secretion into the extracellular space. Thus, it is tempting to speculate that the angiogenesis-promoting effects are executed by xCT and that inhibition of xCT reduces these effects. Therefore, these findings give a mechanistic explanation for the recently reported observation of angiogenesis-promoting effect of glutamate.38
Thus, our data revealed that ATF4 plays a key role in glioma malignancy features and many effects of ATF4 can be pinpointed to the regulation of xCT.
In conclusion, our results demonstrate that ATF4 is elevated in human gliomas and ATF4-induced proliferation can be attenuated by pharmacological or genetic xCT inhibition. Further, ATF4 promotes tumor-mediated neurotoxicity and tumor angiogenesis, which can be alleviated by xCT inhibition and ferroptosis inducers erastin and RSL3. Therefore, ATF4 represents a vital and promising target for anti-tumor strategies and ferroptosis induction in cancer cells.
Concerning the relevance of ATF4-induced chemo-resistancy a study from our laboratory was recently accepted for publication. In this study we provide clinical evidence that increased ATF4 can induce temozolomide (Temcat, Temodal) resistancy: Chen D, Rauh M, Buchfelder M, Eyupoglu IY, Savaskan N. The oxido-metabolic driver ATF4 enhances temozolamide chemo-resistance in human gliomas. doi:10.18632/oncotarget.17737.
Materials and methods
Cell culture
Human U87 and U251 glioblastoma cells were obtained from ATCC/LGC-2397 (Wesel, Germany). Cells were cultured in Dulbecco’s modified Eagle medium (Biochrom, Berlin, Germany) containing 10% fetal bovine serum (Biochrom, Berlin, Germany) with 100 U/ml penicillin, 100 μg/ml streptomycin (Biochrom, Berlin, Germany) and 1% l-glutamine (Gibco/Invitrogen, Carlsbad, CA, USA). Primary rat neurons from newborn Wistar rat (P1–P4) brains were cultured in serum supplemented (10% fetal bovine serum) MEM medium, and additionally supplemented with 2% B27 agent (Invitrogen, Karlsruhe, Germany). Neurons and cell lines were cultured at 37 °C with 5% CO2 and saturated humidified.
Chemicals and reagents
Erastin was purchased from Hycultec GmbH (Beutelsbach, Germany). Sorafenib was purchased from LC Laboratories (Woburn, MA, USA). (1S, 3R)-RSL3 (RSL3) was purchased from Selleckchem BIOZOL (Eching, Germany). Erastin, Sorafenib and RSL3 were dissolved in DMSO under sterile conditions to concentration of 100 mm. Desferoxamine (DFO) and Ferrostatin-1 (Fer-1), Sulfasalazine (SAS), polyclonal rabbit to Laminin, propidium Iodide (PI), Hoechst 33342 and RNAase were bought from Sigma-Aldrich (Taufkirchen, Germany). Desferoxamine was dissolved in water under sterile conditions to a concentration of 50 mm. Ferrostatin-1 was prepared in 50% DMSO/water under sterile conditions to a final concentration of 50 mm. S-4-Carboxy-phenylglycine was purchased from ACRIS Antibodies (Herford, Germany). Sulfasalazine was dissolved in ammonium hydroxide (400 mm) under sterile conditions at 200 mm. S-4-CPG was solved under sterile conditions in 1 m sodium hydroxide to concentration of 100 mM. Mouse monoclonal to ATF4 antibody (ab50546), rabbit polyclonal to xCT antibody (ab37185) and mouse monoclonal to GAPDH antibody (ab8245) was purchased from Abcam (Cambridge, UK). Polyclonal rabbit to phalloidin, 7-Aminoactinomycin (7-AAD), BODIPY C11 (581/591) and Alexa Fluor IgG secondary antibodies were purchased from Life Technologies (Darmstadt, Germany). Polyclonal rabbit to GFAP was purchased from Dako (Glostrup, Denmark). Mouse monoclonal to β-III-tubulin and secondary antibodies used for Western blotting were obtained from Promega (Madison, WI, USA). Roti-fect was obtained from Roth (Karlsruhe, Germany).
Database analysis
The web-based human cancer microarray database NIH/NCI Rembrandt data portal (formerly hosted at https://rembrandt.nci.nih.gov, now moved to http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-3073/ and Georgetown University at https://gdoc.georgetown.edu/gdoc/) and Oncomine (Compendia Biosciences; Ann Arbor, MI, USA; www.oncomine.org) was used to analyze the ATF4 mRNA expression in different malignant brain tissues. Details of the general standardized normalization techniques and statistical calculations can be found on the Oncomine website (https://www.oncomine.com).
Construction of expression plasmids and cell culturing
The full-length complementary DNA (cDNA) of ATF4 and xCT (human ATF4 GenBank accession no. NM_001675.4 and human xCT GenBank accession no. AF252872) were amplified from different DNA templates using a standard PCR-based cloning strategy. PCR products were cloned and ligated into the BgIII-EcoRI site of the peGFP-N1 vector and the peGFP-C1 (Takara, Heidelberg, Germany), respectively. Cells were transfected with ATF4 or xCT expression plasmid using Roti-fect according to the manufacture’s instruction. After 5 days, the culture media was refreshed containing 700 μg/ml Geneticin sulfate 418 (G418; Sigma, St.Louis, MO, USA) for antibiotic selection. After 3 weeks of culturing the selected antibiotics, survival-transfected cells were collected. Stabled-transfected cells were used maintained in 500 μg/ml Geneticin sulfate 418.
Construction of shRNA plasmids and cell transfection
Off-target effects of RNAi were minimized by choosing three 19-mer short interfering RNAs targeting the human ATF4 gene, xCT gene according to the critera of Ui-Tei et al. The selected ATF4 shRNA, xCT shRNA and a scrambled sequence with no homology to any known human genes were synthesized and ligated into the BglII/HindIII sites of pSuperGFP-neo plasmid (OligoEngine) according to the manufacturer’s instruction. ATF4 shRNA, xCT shRNA and srambled shRNA were introduced into glioma cells by transfection with Rofi fect. Cells stably depleted of ATF4 or xCT were obtained by selection with 700 μg/ml G418 as described above.
Cell viability assays and inhibitor studies
Cells were seeded onto 96-well plates (8000 cells per 100 μl per well) and were allowed to attach overnight. For inhibitor studies, cells were treated with indicated concentration for sorafenib, erastin or RSL3 after plating for 12 h. Cell viability was assessed at different time points after treatment (usually 72 h, unless stated otherwise) using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium-bromide (MTT, 5 mg/ml in phosphate-buffered saline; Roth, Karlsruhe, Germany) for 2–3 h. Afterwards, the formazan crystals were then lysed with 100 μl acidic isopropanol and the absorbance of the obtained solutions was determined at 570 nm using TECAN F50 (Crailsheim, Germany) software.
Colony-forming assay
Glioma cells were suspended in 0.5% agar and seeded at ~1000 cells per well of a six-well plate on a preformed 1% agar layer as described in detail.39 Cells were cultured at 37 °C in 5% humidified CO2 for 3 weeks and then washed twice with PBS, methanol-fixed for 15 min, and stained with 0.5% crystal violet for 15 min at room temperature. The colony is defined to consist of at least 50 cells.
Cell morphology analysis
Glioma cells were seeded at a density of 2500/cm2 in 12-well plates on coverslips and cultured under standard conditions. After 2 days, cells were than fixed with 4% formaldehyde (PFA) and stained with phalloidin (1:500) overnight and counterstained Hoechst 33258 (1:10000). Neurons were identified by immunostaining with beta-III-tubulin (1:500), astrocytes with GFAP (1:500) and counterstained with Hoechst 33258 (1:10 000). Coverslips were mounted on slides with Immu-Mount (Thermo Scientific, Barthesville, OK, USA). For imaging an Axio Observer with the Zen Software (Zeiss, Oberkochen, Germany) was acquired.
Real time RT-PCR analysis
Cells were collected in a 15 tube and suspended in 200 μl PBS. We used High Pure RNA Isolation Kit (Roche, Mannheim, Germany) for total RNA extraction according to the manual’s description. RNA content was quantified by NanoVue™ Plus Spectrophotometer (GE Healthcare, UK). cDNA synthesis was performed with the cDNA DyNAmo Kit ((Thermo Scientific, Germany). RT-PCR was performed with the SYBR Green PCR master mix (Thermo Scientific, Germany). Primers specific for ATF4, sense 5′-GGTTCTCCAGCGACA AGG-3′, antisense 5′-TCTCCAACATCCAATCTGTCC-3′; xCT, sense 5′-CCCAGATATGCATCGTCCTT-3′, antisense 5′-GCAACCATGAAGAGGCATGT-3′; GAPDH, sense 5′-TGCACCACCAACTGCTTAGC-3′, antisense 5′-GGCATGGACTGTGGTCATGA-3′. ATF4, xCT and GAPDH primers for RT-PCR were purchased from metabion international AG (Germany). PCR real time cycling conditions were programmed as such: first step (95 °C, 15 min), cycling step (denaturation 94 °C, 15 s; annealing at 56 °C or 60 °C, 30 s; and finally extension for 72 °C, 30 s × 40 cycles), followed by a melting curve analysis to confirm specificity of the PCR using Light Cycler 480 (Roche, Germany). The CT value was corrected by the corresponding GAPDH control CT values.
Immunoblotting
Cells were lysed with NP 40 buffer containing 5 mM NaF and a protease inhibitor cocktail (Roche, Basel, Switzerland) and homogenized by ultrasound (Bandelin Sonoplus, at 67%) as described before.40 In brief, following 20 min incubation on ice, samples were centrifuged at 8000 r.p.m for 10 min. Supernatants were isolated and protein concentration measured with the NanoVue Plus Spectrophotometer (GE Healthcare, UK). Samples were mixed with loading buffer (4 ×) and NuPAGE Sample Reducing Agent (10 ×) (Invitrogen, CA, USA) and boiled at 96 °C for 8 min. Equal amounts of protein lysates were loaded and separated by 10–12% SDS-NuPage gel (Invitrogen, CA, USA) and electrophoresis was performed in MOPS-buffer, transferred to polyvinylidene difluoride (PVDF) membranes (Roth, Karlsruhe, Germany). Pretreatment of membranes was performed in PBS containing 2% Magic block (containing 10% Top block (Lubio science, Luzern, Switzerland)) for 1 h. The membranes were then hybridized with antibodies against ATF4 (1:1000), xCT (1:1000) and GAPDH (1:2000) in 5% BSA-TBST for 10 h at 4 °C. ECL detection was conducted with HRP-conjugated secondary antibody incubation and ECL plus kit (GE healthcare, Solingen, Germany).
Three dimensional spheroid migration assay
A total of 100 000 cells per spheroid were packaged in 100 μl mixture with 32% methylcellulose solution and 68% culture medium. Cell packages were dropped on the petri dishes and reversely cultured (37 °C, 5% CO2, standard humidified conditions) for 12 h so that the cell packages were located with all the cells aggregate to the top and formed spheroid.25, 39 Afterwards the spheroid was moved to the 48-well plate and kept culturing with medium. After 12 h, the spheroid attached to the bottom of the well and the cells migrate, and the pictures of spheroids were taking on the time points day 1, day 2, day 4 and day 6 by Olympus IX71 microscope, and the cells growth area of the spheroid were measured by Image J software (NIH, Bethesda, USA).
Cell cycle analysis
A total of 200 000 cells per well were seeded in 6-well plate and drug treatment was applied for 24 and 72 h.41 Cells and media supernatants were collected as described before.29 Briefly, cells were washed with PBS and fixed with 70% ethanol solution overnight at 4 °C. Fixed cells were washed twice with PBS solution and afterwards resuspended in Hypotonic lysis buffer (0.1% sodium citrate, 0.1% Triton X-100, 100 μg/ml RNAse) for 20 min. Cell cycle analyses were performed within 2 h after adding 7-AAD (7-aminoactinomycin D) by Flow Cytometer BD FACSCanto II (BD Bioscience, Heidelberg, Germany). Analyses were carried out with FlowJo Software 7.6 (Ashland, OR, USA). A minimum of 10 000 cells were counted and analyzed per condition.
BrdU incorporation assay
For BrdU incorporation assay, cells were exposed to medium containing 10 μm 5-bromo-2′-deoxyuridine (BrdU) for 30 min. The cells were then collected, washed to remove unbound BrdU and fixed in 70% ethanol for 45 min on ice. Subsequently, cells were permeabilized with 2 n HCl/Triton X-100 to denature the DNA for 30 min, and incubated with 100 μg/ml RNAse for 30 min. Secondary Alexa Fluor 647-conjugated IgG was added for 30 min at 37 °C, followed by counterstain with 7-AAD. Cellular fluorescence was measured by Flow Cytometer BD FACSCanto II (BD Bioscience). Analyses were carried out with FlowJo Software 7.6.
Cell death analysis by 7-AAD and Annexin V assay
Drugs were applied for 24 and 72 h. After treatment, the Annexin V/7-AAD was assayed (BD Biosciences, Germany) for cell death analysis. Briefly, fresh cell preparations were incubated with 1X Annexin binding buffer and Annexin V/PE (5 μl)-conjugated primary antibody and 7-AAD (5 μl) for 15 min on ice. After incubation, cells were transferred to FACS tubes and analyzed by Flow Cytometer BD FACSCanto II (BD Bioscience, Heidelberg, Germany). A minimum of 10 000 cells were counted and analyzed per condition. Analyses were carried out with FCS Express 5 Demo Software.
Staining for lipid peroxidation in cells by flow cytometry
200 000 cells/well were seeded in 6-well plate. The next day, culture media was replaced with media containing sorafenib (10 μM), erastin (10 μM) or RSL3 (0.1 μM)+/−deferoxamine (DFO, 100 μM) and+/−ferrostatin-1 (Fer-1, 0.5 μM) for 18 h (RSL3, 8 h). After incubation, cells were harvested by trypsinization, washed and resuspended in 500 μl PBS containing BODIPY C11 (2 μM) in FACS tubes. Flow Cytometer BD FACSCanto II was used for the follow cytometer analysis. A minimum of 10 000 cells were counted and analyzed per condition. Analyses were carried out with FCS Express 5 Demo Software.
Metabolic profiling of tumor cells
Unless indicated, all cultures contained 10 mM glucose and 2–4 mm glutamine. To measure consumption and secretion of amino acids from glioma cells, cells were incubated in DMEM without glutamine for 24 h.21 The supernatants were collected and measured by using high performance liquid chromatography (HPLC). Profiles of amino acids were performed by ion-exchange chromatography and post-column ninhydrin derivatization technique using Biochrom 30+ (Laborservice Onken, Gründau, Germany). In brief, 100 μl of each sample was deproteinized with 100 μl of 10% sulphosalicylicacids and 20 μl supernatant was then loaded into a cation-exchange resin-filled column.
VOGIM assay and angiogenesis analysis
Organotypic brain slice cultures were conducted with 5-days-old Wistar rats. Tissues were prepared and maintained as previously described.24, 42 In brief, at day 0, brains were prepared and isolated under ice-cold conditions. Cerebrum was cut into 350 μm thick horizontal slices on a Leica VT 1000S vibratome (Bensheim, Germany) and brain slices were thereafter transferred onto culture plate insert membrane dishes with a pore size of 0.4 μm (GreinerBioOne, Frickenhausen, Germany). Slices were cultured in six-well culture dishes (GreinerBioOne) containing 1.2 ml culture medium (MEM-based HBSS medium, for details see24, 42). The slices were cultured in humidified atmosphere (35 °C, 5% CO2) and washed with 1.2 ml PBS and a full culture medium exchange was performed. At day two, 0.1 μl of the cell-medium-suspension (containing 10 000 con U87, ATF4KD U87 or ATF4OE U87 cells) was placed onto the temporal cortex of the slice by using a 1-μl-Hamilton-syringe. The medium was regularly changed every other day over a course of 7 days. Propidium Iodid (PI) (Sigma) staining was performed on day 5, incubating slices with 1 μg/ml PI for 15–20 min and then wash slices and 6-well plates with PBS. After washing with PBS, complete medium exchange followed. For induction of ferroptosis, brain slices were treated with media containing erastin (10 μm), RSL3 (5 μm)+/−Fer-1 (0.5 μm) for 5 days. On day 7, all the slices were fixed with Immunfixative (4% PFA and picric acid) and stained with anti-laminin-antibody (1:250) for vessel analysis as detailed in Fan et al.38 The number of tumor vessels was quantified by counting the number of vessels which grew into the tumor as described previously.24
Statistical analysis
The data are expressed as mean±s.d. taken from at least three independent experiments, and compared with unpaired Student t-test or Mann–Whitney test where appropriate using SPSS statistics 17.0 software (IBM, Armonk, NY, USA). P-values of P<0.05, **P<0.01; ***P<0.001 were considered as statistically significant.
Patient samples
We used surgical specimens in this study from patients suffering from glioblastoma multiforme (WHO°IV), anaplastic astrocytoma (WHO°III), diffuse astrocytoma (WHO°II) or juvenile pilocytic astrocytoma (WHO°I). Histological diagnoses were performed according to the WHO classification by two independent and experienced neuropathologist (led by Professor Rolf Buslei, Klinikum Bamberg, Germany). Human neocortex specimens negative for human brain tumors served as controls. Patients underwent surgical treatment in the Department of Neurosurgery of the University of Erlangen-Nürnberg, Germany.
Ethical statements
We performed human tissue studies in compliance with the Helsinki Declaration and approved by the Ethics Committee of the Friedrich-Alexander University of Erlangen-Nuremberg. All patients gave written informed consent to participate in the study.
In accordance with the German Protection of Animals Act 14 paragraph, we performed animal killing. These experiments were approved by the designee for animal protection of the University of Erlangen-Nürnberg (TS-7/12). Animal experiments were done in congruence with the EU guidelines for the use of laboratory animals and approved by the Government of Central Franconia (permission to NES).
Accession codes
References
Mao XY, Tokay T, Zhou HH, Jin WL . Long-range and short-range tumor-stroma networks synergistically contribute to tumor-associated epilepsy. Oncotarget 2016; 7: 33451–33460.
Eyupoglu IY, Buchfelder M, Savaskan NE . Surgical resection of malignant gliomas-role in optimizing patient outcome. Nat Rev Neurol 2013; 9: 141–151.
Chevet E, Hetz C, Samali A . Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov 2015; 5: 586–597.
McConkey DJ . The integrated stress response and proteotoxicity in cancer therapy. Biochem Biophys Res Commun 2017; 482: 450–453.
Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM . The integrated stress response. EMBO Rep 2016; 17: 1374–1395.
Singleton DC, Harris AL . Targeting the ATF4 pathway in cancer therapy. Expert Opin Ther Targets 2012; 16: 1189–1202.
Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000; 6: 1099–1108.
Blais JD, Filipenko V, Bi M, Harding HP, Ron D, Koumenis C et al. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol 2004; 24: 7469–7482.
Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 2005; 24: 3470–3481.
Zeng H, Zhang JM, Du Y, Wang J, Ren Y, Li M et al. Crosstalk between ATF4 and MTA1/HDAC1 promotes osteosarcoma progression. Oncotarget 2016; 7: 7329–7342.
Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J 2010; 29: 2082–2096.
Dey S, Sayers CM, Verginadis II, Lehman SL, Cheng Y, Cerniglia GJ et al. ATF4-dependent induction of heme oxygenase 1 prevents anoikis and promotes metastasis. J Clin Invest 2015; 125: 2592–2608.
Fels DR, Koumenis C . The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther 2006; 5: 723–728.
Tanabe M, Izumi H, Ise T, Higuchi S, Yamori T, Yasumoto K et al. Activating transcription factor 4 increases the cisplatin resistance of human cancer cell lines. Cancer Res 2003; 63: 8592–8595.
Igarashi T, Izumi H, Uchiumi T, Nishio K, Arao T, Tanabe M et al. Clock and ATF4 transcription system regulates drug resistance in human cancer cell lines. Oncogene 2007; 26: 4749–4760.
Kilberg MS, Shan J, Su N . ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab 2009; 20: 436–443.
Zhao E, Ding J, Xia Y, Liu M, Ye B, Choi JH et al. KDM4C and ATF4 cooperate in transcriptional control of amino acid metabolism. Cell Rep 2016; 14: 506–519.
Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 2013; 15: 481–490.
Savaskan NE, Heckel A, Hahnen E, Engelhorn T, Doerfler A, Ganslandt O et al. Small interfering RNA-mediated xCT silencing in gliomas inhibits neurodegeneration and alleviates brain edema. Nat Med 2008; 14: 629–632.
Robert SM, Buckingham SC, Campbell SL, Robel S, Holt KT, Ogunrinu-Babarinde T et al. SLC7A11 expression is associated with seizures and predicts poor survival in patients with malignant glioma. Sci Transl Med 2015; 7: 289ra86.
Sehm T, Fan Z, Ghoochani A, Rauh M, Engelhorn T, Minakaki G et al. Sulfasalazine impacts on ferroptotic cell death and alleviates the tumor microenvironment and glioma-induced brain edema. Oncotarget 2016; 7: 36021–36033.
Dixon SJ, Stockwell BR . The role of iron and reactive oxygen species in cell death. Nat Chem Biol 2014; 10: 9–17.
Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 2014; 16: 1180–1191.
Ghoochani A, Yakubov E, Sehm T, Fan Z, Hock S, Buchfelder M et al. A versatile ex vivo technique for assaying tumor angiogenesis and microglia in the brain. Oncotarget 2016; 7: 1838–1853.
Xu T, Fan Z, Li W, Dietel B, Wu Y, Beckmann MW et al. Identification of two novel Chlorotoxin derivatives CA4 and CTX-23 with chemotherapeutic and anti-angiogenic potential. Sci Rep 2016; 6: 19799.
Sehm T, Rauh M, Wiendieck K, Buchfelder M, Eyupoglu IY, Savaskan NE . Temozolomide toxicity operates in a xCT/SLC7a11 dependent manner and is fostered by ferroptosis. Oncotarget 2016; 7: 74630–74647.
Conrad M, Angeli JP, Vandenabeele P, Stockwell BR . Regulated necrosis: disease relevance and therapeutic opportunities. Nat Rev Drug Discov 2016; 15: 348–366.
Iurlaro R, Munoz-Pinedo C . Cell death induced by endoplasmic reticulum stress. FEBS J 2016; 283: 2640–2652.
Savaskan NE, Seufert S, Hauke J, Trankle C, Eyupoglu IY, Hahnen E . Dissection of mitogenic and neurodegenerative actions of cystine and glutamate in malignant gliomas. Oncogene 2011; 30: 43–53.
Takeuchi S, Wada K, Toyooka T, Shinomiya N, Shimazaki H, Nakanishi K et al. Increased xCT expression correlates with tumor invasion and outcome in patients with glioblastomas. Neurosurgery 2013; 72: 33–41.
Timmerman LA, Holton T, Yuneva M, Louie RJ, Padro M, Daemen A et al. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell. 2013; 24: 450–465.
Savaskan NE, Fan Z, Broggini T, Buchfelder M, Eyupoglu IY . Neurodegeneration and the Brain Tumor Microenvironment. [corrected]. Curr Neuropharmacol 2015; 13: 258–265.
Savaskan NE, Eyupoglu IY . xCT modulation in gliomas: relevance to energy metabolism and tumor microenvironment normalization. Ann Anat 2010; 192: 309–313.
Wang Y, Ning Y, Alam GN, Jankowski BM, Dong Z, Nor JE et al. Amino acid deprivation promotes tumor angiogenesis through the GCN2/ATF4 pathway. Neoplasia 2013; 15: 989–997.
Castranova D, Davis AE, Lo BD, Miller MF, Paukstelis PJ, Swift MR et al. Aminoacyl-transfer RNA synthetase deficiency promotes angiogenesis via the unfolded protein response pathway. Arterioscler Thromb Vasc Biol 2016; 36: 655–662.
Chakraborty G, Jain S, Kundu GC . Osteopontin promotes vascular endothelial growth factor-dependent breast tumor growth and angiogenesis via autocrine and paracrine mechanisms. Cancer Res 2008; 68: 152–161.
Pereira ER, Frudd K, Awad W, Hendershot LM . Endoplasmic reticulum (ER) stress and hypoxia response pathways interact to potentiate hypoxia-inducible factor 1 (HIF-1) transcriptional activity on targets like vascular endothelial growth factor (VEGF). J Biol Chem 2014; 289: 3352–3364.
Fan Z, Sehm T, Rauh M, Buchfelder M, Eyupoglu IY, Savaskan NE . Dexamethasone alleviates tumor-associated brain damage and angiogenesis. PLoS One 2014; 9: e93264.
Fan Z, Bittermann-Rummel P, Yakubov E, Chen D, Broggini T, Sehm T et al. PRG3 induces Ras-dependent oncogenic cooperation in gliomas. Oncotarget 2016; 7: 26692–26708.
Broggini T, Schnell L, Ghoochani A, Mateos JM, Buchfelder M, Wiendieck K et al. Plasticity Related Gene 3 (PRG3) overcomes myelin-associated growth inhibition and promotes functional recovery after spinal cord injury. Aging (Albany NY) 2016; 8: 2463–2487.
Ghoochani A, Schwarz MA, Yakubov E, Engelhorn T, Doerfler A, Buchfelder M et al. MIF-CD74 signaling impedes microglial M1 polarization and facilitates brain tumorigenesis. Oncogene 2016; 35: 6246–6261.
Ghoochani A, Hatipoglu Majernik G, Sehm T, Wach S, Buchfelder M, Taubert H et al. Cabazitaxel operates anti-metastatic and cytotoxic via apoptosis induction and stalls brain tumor angiogenesis. Oncotarget 2016; 7: 38306–38318.
Acknowledgements
We thank all members of the Translational Neurooncology laboratory for valuable comments on this study and helpful suggestions. Christiane Mühle (Department of Psychiatry, Erlangen, Germany) is gratefully acknowledged for providing assistance with the digital qRT-PCR procedure. Christoph J. Wruck (from the RWTH Aachen, Germany) as well as Jana Wrosch and Marc Dahlmanns (from the Department of Psychiatry, Erlangen, Germany, and Molecular Medicine program at the Universitätsklinikum Erlangen) are acknowledged for sharing imaging technics and constructs exchange. DC holds a grant from the China Scholarship Council (CSC: 201308110471). This study was funded by the Verein zur Förderung des Tumorzentrums der Universität Erlangen-Nürnberg e.V. to NES.
Author contributions
NES and DC designed and conceived the study. NES supervised the study. DC performed the in vitro experiments with help from ZF, MR and DC performed the AA analysis. The data analysis and evaluation was conducted by DC and ZF and critically discussed with all authors. NES wrote the manuscript in conjunction with DC. All authors contributed to the preparation of the final manuscript and lent shape to the final paper. DC accomplished the present work in fulfillment of the requirements for obtaining the degree Dr. Med. at the Medical Faculty of the Friedrich-Alexander University of Erlangen-Nürnberg (FAU).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Chen, D., Fan, Z., Rauh, M. et al. ATF4 promotes angiogenesis and neuronal cell death and confers ferroptosis in a xCT-dependent manner. Oncogene 36, 5593–5608 (2017). https://doi.org/10.1038/onc.2017.146
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.146
This article is cited by
-
Emerging mechanisms of the unfolded protein response in therapeutic resistance: from chemotherapy to Immunotherapy
Cell Communication and Signaling (2024)
-
Augmented ERO1α upon mTORC1 activation induces ferroptosis resistance and tumor progression via upregulation of SLC7A11
Journal of Experimental & Clinical Cancer Research (2024)
-
Targeting paraptosis in cancer: opportunities and challenges
Cancer Gene Therapy (2024)
-
Ferroptosis: Emerging mechanisms, biological function, and therapeutic potential in cancer and inflammation
Cell Death Discovery (2024)
-
Targeting ferroptosis in hepatocellular carcinoma
Hepatology International (2024)