Abstract
Chronic inflammation (CI) is a risk factor for pancreatic cancer (PC) including the most common type, ductal adenocarcinoma (PDAC), but its role and the mechanisms involved are unclear. To investigate the role of CI in PC, we generated genetic mouse models with pancreatic specific CI in the presence or absence of TP53. Mice were engineered to express either cyclooxygenase-2 (COX-2) or IκB kinase-2 (IKK2), and TP53+/+ or TP53f/f specifically in adult pancreatic acinar cells by using a full-length pancreatic elastase promoter-driven Cre. Animals were followed for >80 weeks and pancreatic lesions were evaluated histologically and immunohistochemically. The presence of K-ras mutations was assessed by direct sequencing, locked nuclei acid (LNA)-based PCR, and immunohistochemistry. We observed that sustained COX-2/IKK2 expression caused histological abnormalities of pancreas, including increased immune cell infiltration, proliferation rate and DNA damage. A minority of animals with CI developed pre-neoplastic lesions, but cancer was not observed in any TP53+/+ animals within 84 weeks. In contrast, all animals with CI-lacking TP53 developed various subtypes of PC, including acinar cell carcinoma, ductal adenocarcinoma, sarcomatoid carcinoma and neuroendocrine tumors, and all died within 65 weeks. No evidence of K-ras mutations was observed. Variations in the activity of the Hippo, pERK and c-Myc pathways were found in the diverse cancer subtypes. In summary, chronic inflammation is extremely inefficient at inducing PC in the presence of TP53. However, in the absence of TP53, CI leads to the development of several rare K-ras-independent forms of PC, with infrequent PDAC. This may help explain the rarity of PDAC in persons with chronic inflammatory conditions.
Similar content being viewed by others
Introduction
The link between inflammation and cancer, first suggested in the nineteenth century by Rudolf Virchow, who observed that tumors are frequently infiltrated with immune cells, is now accepted as an important component of cancer development.1 The risk of developing pancreatic cancer (PC) is elevated by a variety of inflammatory conditions including chronic pancreatitis, alcohol abuse, smoking and obesity.2 Chronic inflammation (CI) is also associated with several other cancers including gastric, colorectal and liver.2, 3 However, most people with chronic inflammatory conditions do not develop cancer, suggesting that chronic inflammation is not efficient at driving carcinogenesis. Nonetheless, inflammation can directly affect epithelial cells to generate reactive oxygen and nitrogen species that lead to DNA damage and result in genetic instability.4 Genetic instability increases the probability of random genetic alterations some of which are carcinogenic.5 Currently, the consequences of prolonged inflammation in the pancreas are unclear, as are the mechanisms responsible for protecting the cells from the potentially carcinogenic effects of inflammation.
In the current study, we utilized genetic models expressing clinically relevant genes to generate chronic inflammation in the pancreas. Mice were developed with pancreatic expression of either cyclooxygenase-2 (COX-2) or IĸB kinase-2 (IKK2), in the presence or absence of TP53. COX-2 or IKK2 activity, which is often observed in patients with inflammation, results in generation of a pathologically relevant variety of mediators. In the current study, specific expression or deletion relevant genes were achieved by crossing floxed or floxed-stopped versions of these molecules with mice bearing a Cre recombinase driven by a full-length mouse pancreatic elastase promoter previously found to be highly efficient and specific for adult pancreatic acinar cells6 (Supplementary Figure 1). The mice were observed for periods of up to 84 weeks and the progression of pancreatic changes was assessed.
Surprisingly, no tumors were observed with either driver of CI when TP53 was present. However, in the absence of TP53, complex tumors with multiple subtypes of K-ras-independent cancers were always observed. These data suggest that initial loss of TP53 may be important for the development of rare forms of pancreatic cancer. Evidence that genetic instability was responsible for carcinogenesis caused by CI in the absence of TP53 included the long latency required for tumor formation, the direct detection of DNA damage, and the complexity of the resulting tumors. These findings help explain the low frequency of pancreatic ductal adenocarcinoma (PDAC) in patients with chronic inflammatory conditions and the rarity of non-PDAC tumors in the pancreas.
Results
Targeted expression in pancreas of COX-2 and/or IKK2 leads to chronic inflammation but does not readily induce tumorigenesis in the presence of wild-type TP53
Analysis of pancreatic histology revealed slowly progressive chronic inflammation in mice expressing COX-2 (Figure 1a). In the majority of COX-2 expressing mice, the pancreata did not show significant pathological changes at 20 weeks of age, but by weeks 40 and 80, prominent acinar atrophy, vacuolization and moderate levels of acinar to ductal metaplasia (ADM) were evident. At later times, ductal proliferation and ectasia were the most prominent features. Low frequencies of early pancreatic intraepithelial neoplasia (PanIN) lesions ((PanIN1 in 35% (7/20); PanIN2 in 5% (1/20)) were observed. (Figure 1a). COX-2 expression caused increased expression of proinflammatory and profibrogenic cytokines (Supplementary Figure 1D). The presence of CI was confirmed by increased immune cell infiltrate including leukocytes and pancreatic stellate cells, with macrophages being the most common. A slow accumulation of collagen deposition was also observed (Figure 1a; Supplementary Figures 2A and B). The progression of changes was associated with an increase in the proliferation rate assessed by Ki-67 staining, with the highest degree of proliferation within ducts (Supplementary Figures 2C and D; Figure 1c). DNA damage was also evident in mice expressing COX-2, as indicated by increased cells with nuclei positive for γH2AX foci (Figure 1c; Supplementary Figure 2E).
Pancreas-specific COX-2 and IKK2 overexpression caused inflammatory changes but were not sufficient to generate pancreatic tumors. (a) Representative H&E images showing progressive chronic inflammatory changes in COX-2/Cre mice at 20, 40 and 80 weeks, including atrophy of acinar compartment, acinar to ductal metaplasia and pancreatic intraepithelial neoplasia (n=10 per group; scale bar: 200 μm, magnification 10 ×). The immunohistochemistry showing the progression of changes associated with the increase in inflammatory cell infiltrate (leukocytes, CD45; macrophages: F4/80; pancreatic stellate cells: α-SMA) (scale bar: 200 μm, magnification 20 ×), and fibrosis (Picro-Sirius Red) (scale bar: 200 μm, magnification 10 ×). (b) Representative H&E images showing changes in IKK/Cre mice at 20, 40 and 80 weeks including atrophy of acinar compartment and fatty replacement, and immunohistochemistry for inflammatory cell infiltrates (leukocytes: CD45; macrophages: F4/80) (scale bar: 200 μm, magnification 20 ×), and fibrosis (scale bar: 200 μm, magnification 10 ×). (c) The immunohistochemistry for γH2AX and proliferation rate (Ki-67) in COX-2/Cre, IKK/Cre and control (TP53+/+) mice (scale bar: 100 μm, magnification 40 ×). (d) Representative H&E images of pancreas of TP53−/−/Cre and control (TP53+/+) mice at the age of 80 weeks (scale bar: 200 μm, magnification 10 ×).
The histologic changes in the pancreata of IKK2 expressing mice were generally similar to those of COX-2 expressing mice. Both IKK2 and COX-2 expression caused progressive loss of acinar cells (Figures 1a and b). IKK2 expression induced the infiltration of leukocytes, indicating ongoing inflammation, and increased proliferation rate and DNA damage (Figures 1b and c; Supplementary Figures 2A and C–E). However, unlike COX-2 mice, IKK2 bearing mice showed no evidence of ADM and the most prominent histologic change was fatty replacement of the pancreas. In addition, IKK2 bearing mice showed no increase in pancreatic fibrosis (Figure 1b). Despite prominent chronic inflammatory changes, neither the expression of COX-2 nor IKK2 nor the expression of both COX-2 and IKK2 led to PC development in any mice during the observation period of 84 weeks.
Chronic inflammation leads to multiple subtypes of pancreatic cancer in the absence of TP53
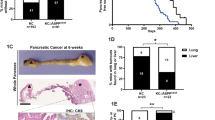
Deletion of TP53 in the pancreas had no obvious effects on pancreatic histology, nor did any TP53−/−/Cre mice developed pancreatic tumors (Figure 1d). However, when COX-2/IKK2 were expressed in mice lacking TP53, all mice developed abdominal swelling, a palpable mass, became moribund, or showed continuous weight loss, leading to death or humanitarian sacrifice. All of the mouse models with CI and TP53 deletion had significantly shorter survival times compared with those only lacking TP53. None of the mice with CI-lacking TP53 survived longer than 65 weeks (Figure 2A). However, we did not observe statistically significant differences among the TP53−/−/COX-2/Cre, TP53−/−/IKK/Cre and TP53−/−/COX-2/IKK/Cre mice.
Lack of TP53 and overexpressing COX-2 and/or IKK2 significantly reduced survival and led to the development of mixed pancreatic carcinomas. (A) Mice overexpressing COX-2 and/or IKK2 with TP53 deletion had significantly shorter survival compared with the mice with TP53 deletion alone and controls (TP53+/+). Note: long median survival (Mantel–Cox test) of all groups. (B) Representative H&E images showing histological heterogeneity of tumors; a,b. PDAC: Less cellular, abundant fibrous stroma; (c–f). ACC: highly cellular with scant fibrous stroma and frequent necrosis; (d–f). ACC histological patterns: trabecular (c); acinar (d), solid (e and f); acinar cell cystadenoma (g); sarcomatoid carcinoma (h); chronic inflammatory changes associated with tumor (i) (scale bar: 200 μm, magnification: 10 ×, 20 ×). (C) Immunohistochemistry for CK-19, amylase, synaptophysin and chromogranin A confirmed the mixed histology of tumors. (D) Loss of TP53 function in combination with COX-2/IKK overexpression leads to metastatic pancreatic carcinoma. (a, b) Representative H&E of liver metastases. (c) Positive immunohistochemistry for amylase indicate acinar cell carcinoma origin of metastases. (d) Non-specific immunohistochemistry for CK-19 (scale bar: 200 μm, magnification: 10 ×).
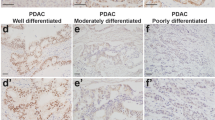
The mice with CI and TP53 deletion developed large pancreatic tumors up to 3 cm (Supplementary Figure 3A). Examination of these tumors revealed mixed carcinomas of different histologic subtypes, including acinar cell carcinoma (ACC), ductal adenocarcinoma (PDAC), poorly differentiated tumors with sarcomatoid features (SC), as well as neuroendocrine carcinoma (NEC) (Figures 2B and C and Table 1). Interestingly, PDAC, while sometimes present, was not the main component of the mixed carcinomas (30% mice, 6/20) and was always observed mixed with ACC, SC and/or NEC in these mice. For definitive identification of each histologic subtypes, we performed immunohistochemical staining. PDAC was diagnosed as area of tumor showing the typical histological features of ductal adenocarcinoma and positive staining for CK-19 (Figure 2C), a marker of ductal differentiation. ACC was identified as a region with acinar or solid growth pattern showing a positive immunoreactivity for amylase and the absence of cytokeratin 19 staining (Figure 2C). NEC was identified using neuroendocrine markers chromogranin A and synaptophysin. In some tumors, the neuroendocrine carcinoma compartment represented more than 25% of neoplastic cells, which fits the diagnostic criteria for mixed carcinomas of pancreas in humans (Figure 2C).7, 8
Variability in the immune cell infiltration was also observed within tumors (Supplementary Figures 3B and C). Among the variety of cells infiltrating the tumor, we detected an increase in the number of cells positive for Stem cell antigen-1 (Sca-1) (Supplementary Figure 3C). Sca-1 is widely used to detect hematopoietic stem cells and is a candidate marker in the search for tissue-resident and cancer stem cells. Sca-1 is upregulated in many tumors such as retinoblastoma, mammary gland and prostate cancer. Fibroblastic tumors and mammary adenocarcinoma cells expressing high Sca-1 levels are more malignant than tumors with low Sca-1 expression.9 In addition, we detected the presence of single cells Sca-1 positive in COX-2/Cre and IKK/CreBAC mice (Supplementary Figure 3D). We also assessed the proliferation rate in tumors, which was significantly higher compared with controls, with variability observed between tumor types (Supplementary Figures 4A–D). We also observed DNA damage by positive immunohistochemistry for γH2AX (Supplementary Figures 4E and F). It has been previously reported, that TP53 deletion contributes to metastasis of PC.10 In this study, we observed metastases to the liver in 30% of tumors. Surprisingly, most of the metastatic carcinomas showed positive strong immunoreactivity for amylase, and weak or negative staining of CK-19, which were consistent with ACC (Figure 2D).
Cancers induced by chronic inflammation in the absence of TP53 showed heterogeneity of cancer-related genes and pathways but oncogenic mutations of K-ras were not detected
To further characterize the tumors in our mouse models, we next analyzed for the presence or absence of the common genetic alterations known in PC. Point mutations (G12D, G12V) in K-ras are present in the majority of PDAC. In the current study, direct DNA sequencing did not detect K-ras mutations in microdissected histologically defined PDAC areas. It is known to be very difficult to accurately detect the scarce copies of single-base mutated genes in neoplastic tissues among thousands of copies of wild-type DNA. Therefore, we further examined the samples using an alternative approach to analyze gDNA using an locked nuclei acid (LNA) probe to suppress the amplification of the WT allele, which was previously described as allowing sensitive detection of mutant K-ras.11 Consistent with our direct DNA sequencing results, we did not reveal the presence of K-ras mutations using this approach (Figure 3A). In addition, immunohistochemistry using an antibody specific for mutant K-ras did not provide positive staining in these tumors, though it readily identified mutant K-ras expressing cells in control LSL-Kras mice (Figure 3B; Supplementary Figure 5). Despite the lack of detection of mutant K-Ras, the expression of pERK, which can act as a marker of Ras activation was observed in scattered cells in the areas of PDAC (Figure 3C), as well as occasionally in the expanded ducts in COX-2 expressing animals and acinar cells in IKK expressing mice (Supplementary Figure 6) but was not observed in control mice or non-PDAC tumors.
Ras-independent tumorigenesis was the predominant pathway of carcinogenesis associated with chronic inflammation in the absence of TP53. (A) Detection of K-ras mutation. (a) In control mice, only wild-type (WT) DNA (negative control) signal was detected in absence of LNA probe, whereas addition of LNA probe resulted in its complete suppression. (b) In K-RasG12D expressing mice (LSLG12D/Cre) adding the LNA probe resulted in suppression of WT signal and detection of mutant K-ras as indicated by the difference in the melting temperature for WT DNA and mutant K-ras. (c) The results of gDNA analysis of microdissected tissue from PDAC areas of tumor showing detection of only WT but not mutant K-ras. (B) Representative Immunohistochemical images showing for mutant Ras showing positive staining in LSL-K-rasG12V/Cre and LSL-K-rasG12D/Cre tumors, and negative staining in tumors of TP53−/−/COX-2/Cre mice (scale bar: 200 μm, magnification 20 ×). (C) Representative immunohistochemical images showing focal positive staining for phospho-ERK (pERK), in tumors (two representative images of TP53−/−/COX-2/Cre and TP53−/−/COX-2/IKK/Cre tumors) (scale bar: 200 μm, magnification 10 ×).
The tumor suppressor p16 is lost in around 50% of PDAC. However, in genetic mouse models it has been observed that tumors generated by the expression of oncogenic K-ras and deletion of TP53 typically retain p16.12 In agreement with these previous observations, in the present study we found persistent expression of p16 in the majority of the tumors, including within the limited PDAC component. However, loss of p16 expression was observed in some areas of poorly differentiated carcinoma with sarcomatoid features (Figure 4; Supplementary Figure 7), which may indicate more extensive genetic alterations in this cancer subtype.
Persistent expression of p16 in majority of tumors, including PDAC areas and loss of its expression in undifferentiated tumors. Representative immunohistochemical images showing p16 expression in control (TP53+/+), TP53−/−/Cre, COX-2/Cre and TP53−/−/COX-2/Cre mice (scale bar: 200 μm, magnification: 20 ×).
There is growing evidence that the Hippo signaling pathway may have an important role in PC.13 We noted that ectopic expression of COX-2 led to the upregulation of the major Hippo pathway molecule, Yes-associated Protein (YAP). YAP had a predominant nuclear localization (indicating activation of Hippo pathway) in ductal cells of COX-2/Cre mice at the age of 20 weeks and an increase in nuclear expression mainly within ADM and PanIN lesions at later stages (Supplementary Figure 8A). We determined the YAP expression in the mixed carcinomas developed in this model. The PDAC component showed a predominant nuclear expression of YAP, similar to what is observed in oncogenic K-ras models (LSL-KrasG12D/Cre mice) (Supplementary Figures 8A and B). In contrast, non-PDAC areas of tumors showed predominant cytoplasmic YAP localization and sarcomatoid tumors showed complete loss of YAP expression. In addition, we detected nuclear YAP expression in stromal cells associated with tumors (Figure 5; Supplementary Figure 8A). Our findings confirmed the important role of Hippo pathway in pancreatic tumorigenesis and indicate that different YAP expression patterns occur in different pancreatic tumor types.
Immunohistochemical heterogeneity of tumors for pERK, YAP and c-Myc. (a) PDAC areas of tumors showed positive CK-19 and pERK expression, and nuclear expression of YAP and pS62c-Myc. Non-PDAC areas of tumors showed negative CK-19 and pERK expression, and negative or cytoplasmic YAP expression (scale bar: 200 μm, magnification: 20 ×). (b) The calculation of the % expression of pERK (n=7), c-Myc (n=5) and YAP (n=5) in PDAC and non-PDAC areas (***P<0.001).
Levels of activated c-Myc and Akt were also evaluated. c-Myc protein has an essential role in DNA damage-induced apoptosis through the control of TP53 protein.14 Interestingly, c-Myc induced transformation of acinar cells in transgenic mouse was previously shown to produce both acinar and mixed acinar/ductal neoplasms.15 More recently, serine 62 phosphorylated Myc (pS62Myc), a key residue that is phosphorylated in response to mitogen stimulation and Ras-raf-MEK-ERK signaling, has been shown to play a role in the induction of proliferation of many cells and to be essential for regenerative proliferation in response to DNA damage.16 We found increased nuclear expression pS62Myc only in the PDAC component of the mixed carcinomas and in stromal cells (Figure 5). Increased expression was also detected within ADM and PanIN lesions in COX-2/Cre mice (Supplementary Figure 9A). PI3K/Akt signaling is one of the most commonly deregulated signaling pathways in cancer, including PDAC.17 We detected increased pAkt expression within ADM and PanIN lesions in mice overexpressing COX-2 while only focal pAkt expression was found within tumors (Supplementary Figure 9B).
Discussion
In this study, chronic inflammation of the pancreas induced by expression of either COX-2 or IKK2 caused major pathological alterations, but did not induce PC in animals with WT TP53 within 84 weeks (1.6 years). This indicates that chronic inflammation alone is an inefficient driver of PC. In contrast, in the absence of TP53, mixed carcinomas consisting of multiple pancreatic cancer subtypes including PDAC, ACC, SC and NEC consistently developed and were fatal within 65 weeks. Surprisingly, PDAC was not the predominant component in these tumors and no K-ras mutations were observed. Rather, the histological heterogeneity of the tumors was accompanied by heterogeneous alterations in cancer-related pathways including YAP, pERK, c-Myc and Akt. Therefore, CI, even in the absence of TP53, is ineffective at inducing PDAC, the most common type of human PC. These observations may help explain the rare occurrence of pancreatic cancer in patients with CP and other inflammatory conditions.
In the current study, CI was achieved by transgenic expression of COX-2 or IKK2 in adult pancreatic acinar cells. IKK2 leads to the continuous phosphorylation and degradation of inhibitory proteins controlling NF-kB activation.18 NF-kB activity is high in CP and PDAC where it regulates the expression of genes involved in inflammation and cell survival.19 COX-2 is also upregulated in inflammation and is high in patients with CP and PDAC. COX-2 activation is associated with the generation of inflammatory molecules including prostaglandins, prostacyclin and thromboxane A2.20, 21, 22 Previous genetic mouse models with ectopic expression of COX-2 driven either from the CK5 or elastase promoter reported development of pancreatic fibrosis and a variety of premalignant alterations but no metastatic tumors.23, 24 Interestingly, expression of COX-2 from the CK5 promoter, which targeted duct cells, caused severe histological alterations and itself was lethal within ~30 weeks.23 However, it is unclear whether the deaths in that mouse model were accounted for by expression within the pancreas or at other sites. In another study, when inflammatory cytokine IL1b was expressed in mice bearing a copy of mutant TP53R172H/+ there was an increase in ADM, but again no tumors were observed.25 In the current study, expression of COX-2 or IKK2 in adult acinar cells did not result in tumors within 84 weeks. However, the presence of PanIN lesions in a small minority of the COX-2 expressing mice suggests that with more time cancer might develop and would likely be PDAC.
In the current study, chronic inflammation led to DNA damage in pancreatic acinar cells. In the presence of TP53, this DNA damage caused the loss of acinar cells. The most likely explanation for the observations made in this study is that, in the absence of TP53 mediated apoptosis or senescence, the genetic instability5 caused by CI eventually led to carcinogenesis. The surviving genetically unstable cells developed of a variety of cancer subtypes, indicating no favored pathway to transformation (Figure 6). The development of cancers in the absence of TP53 required a long latency of at least ~9 months. This suggests that even with high levels of genetic instability, the probability of developing alterations required for carcinogenesis is low. We were not able to detect the most common K-ras mutations, which are the most frequent genetic alterations found in PDAC.
PDAC often involves oncogenic mutations of K-ras, with the reported frequency of K-ras mutations in studies of patient samples of PDAC varying greatly from 92% to only 47.2%.26, 27 The discrepancies in mutation frequencies could be attributable to differences in the detection methods, specimen types, or the racial characteristics of the patients. In the current study, we were unable to detect the most common mutation of K-ras (G12) even in microdissected areas of PDAC. Single nucleotide mutations are difficult to detect in a background of wild-type genes. Thus, it remains possible that mutations of K-ras existed in some areas of PDAC. We also did not assess other K-ras mutations that are rare in human PDAC samples.28 Nonetheless, these observations indicate that oncogenic mutation of K-ras was clearly not the predominant pathway of carcinogenesis in these studies. Furthermore, these data indicate that K-ras is neither favored nor necessary for the development of the rarer forms of PC such as ACC or endocrine carcinoma. Interestingly, it was previously observed that PDAC occurring in mouse models with genetic instability caused by BRACA2 deletion plus interference with TP53 were reported to develop invasive PDAC of variable histological features with relatively long latency and to have a very low frequencies of secondarily acquired K-ras gene mutations.29, 30 Thus, genetic instability per se does not favor mutation of K-ras but rather leads to a variety of genetic alterations and results in heterogeneous tumors.
In the current study, in the absence of TP53 a high percentage of rare tumors were observed, such as ACC, which accounts for about 5 and 15% of PC in adults and children, respectively.7, 8 ACC is not associated with K-ras mutations.7, 8, 31 Recently, TP53 genetic alterations were found in ~50% of ACC and were correlated with worse survival.32 In the current study, sarcomatoid and neuroendocrine carcinomas were also observed. The preponderance of these rarer forms of PC in these models suggests that loss of TP53 is normally a rare event. Potentially this is explained by the need for two genetic events to lose TP53 function, as a single wild-type allele is sufficient for TP53 activity. The probability of random DNA instability generating two genetic events to block TP53 function is lower than the probability of developing a single mutation in the K-ras gene. This may explain the relative abundance of PDAC compared with the other forms of PC, because when K-ras is mutated as a first hit, the cancer that develops is always PDAC.
In contrast to the relative inefficiency of chronic inflammation to initiate PC, it has been well established that inflammation can accelerate the development of cancer in cells with preexisting oncogene expression. For PC, inflammation greatly accelerates the development of cancer in adult cells bearing activating mutations of K-ras.33 This effect of inflammation in cells expressing endogenous levels of oncogenic K-ras has been partially explained by the observation the mutant K-ras has very low activity until stimulated by external factors such as inflammatory stimuli.34, 35, 36 However, once Ras activity is elevated sufficiently, it generates inflammation through its ability to activate mechanisms such as COX-2 and NF-kB, which can perpetuate the cycle.35 Furthermore, the inflammation generated by high Ras activity is a form of CI and induces genetic instability, increasing the probability of losing a tumor suppressor and developing PDAC. Thus, inflammation is clearly a risk factor once K-ras mutations exist.
In conclusion, this study improves our understanding of the role of chronic inflammation and TP53 loss in the development of pancreatic cancers. The data indicate that chronic inflammation in the presence of TP53 is inefficient at inducing carcinogenesis. In contrast, chronic inflammation in the absence of TP53 generates acinar, ductal, neuroendocrine and sarcomatoid tumors, most of which are rare in patients and are independent of K-ras mutations. Taken together with epidemiological data, these observations suggest that oncogenic mutation of K-ras is a rare consequence of chronic inflammation, but when it occurs it can lead to PDAC. Clearly the mechanisms responsible for K-Ras mutations are important and currently unknown. The loss of TP53 function appears to be even less probable, but when it occurs the result can be several different forms of pancreatic cancer which are K-ras independent.
Materials and methods
Genetically engineered transgenic mice
A full-length elastase (Ela) gene promoter was used to drive the expression of tamoxifen-regulated CreERT specifically in adult pancreatic acinar cells in mice (Ela-CreERT) as described previously.6 COX-2-floxed mice were obtained from Harvey R. Herschman, UCLA, Los Angeles, California, USA.37 For targeted expression of COX-2 in pancreatic acinar cells, COX-2 floxed mice were bred with Ela-CreERT mice to generate COX-2/Ela-CreERT mice (referred as COX-2/Cre; Supplementary Figure 1). Transgenic mice expressing conditionally active IKK2 were developed as described previously.18 Briefly, a fragment containing loxP-GFP-stop-loxP followed by active IKK2 (produced by conversion of 2 conserved serines in the T loop of IKK2 with glutamates; a kind gift from Dr. Michael Karin at the University of California, San Diego, USA) was cloned into a pCAGGS vector (provided by Dr Miyazaki, Kamamoto University Medical School, Japan), which contains a cytomegalovirus and a chicken-actin promoter that generates high levels of expression. All transgenic mice were developed by pronuclear injection. For targeted expression of IKK2 in pancreas, IKK2 mice were bred with Ela-CreERT mice to generate IKK2/Ela-CreERT mice (referred as IKK/Cre). Transgenic expression of GFP was visualized under a UV lamp. The TP53-floxed conditional deletion mice were obtained from the Mouse Models for Human Cancer Consortium Repository (Rockville, MD, USA).38 For targeted pancreas-specific TP53 deletion, mice were bred with Ela-CreERT mice to generate TP53−/−/Cre and with COX-2/Cre and IKK/Cre mice to generate: TP53−/−/COX-2/Cre, TP53−/−/IKK/Cre, and TP53−/−/COX-2/IKK/Cre mice. Tamoxifen treatment was used to induce transgene expression. All experiments were conducted in compliance with the regulations and ethical guidelines for experimental animal studies of the Institutional Animal Care and Use Committee at the University of Texas M.D. Anderson Cancer Center.
Histology
Pancreatic tissues were fixed with 10% formaldehyde in phosphate-buffered saline (PBS), embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E). Histological analysis was performed by pathologists blinded to experimental procedures. Each mouse pancreas was assessed for the presence of pancreatic changes, including inflammatory cell infiltrate, acinar cell atrophy, fibrosis, ADM, PanIN and histologic subtypes of PC.
Immunohistochemistry
Immunohistochemical staining was performed on pancreatic paraffin sections. Briefly, after deparaffinization, antigen retrieval (DAKO target retrieval solution, DAKO, Carpentaria, CA, USA) was performed in a steamer for 20 min at 98 °C. Endogenous peroxidase activity was blocked with H2O2, followed by washing and blocking, and primary antibodies were applied: α-smooth muscle actin (1:200, Abcam, Cambridge, MA, USA, ab5694), amylase (1:400, ab21156, Abcam), chromogranin A (1:200, Santa Cruz Biotechnology, Dallas, TX, USA, sc1488), cleaved caspase-3 (1:100, Cell Signaling Technology—CST, Danvers, MA, USA, 9664), COX-2 (1: 300; Cayman Chemicals, Ann Arbor, MI, USA), CK-19 (1:100, Abcam, ab15463), CD45 (1:500, BD Pharmingen, San Jose, MO, USA, 553078), F4/80 (1:250, e-Bioscience, San Diego, CA, USA, 14-4801), Ki-67 (1:200, Sigma-Aldrich, St Louis, MO, USA, AB9260), pAkt (1:50, CST, 4060), phospho-ERK (1:200, CST, 4370); CDNK2A/p16INK4a (1:2000, Abcam, ab54210), phospho-histone H2A.X (1:100, CST, 9718), pS62c-Myc (1:100, Abcam, ab51156), Ras (G12D) mutant specific (1:100, CST, 14429), YAP (1:500, CST, 14074), YAP (1:250, Santa Cruz Biotechnology, sc 15407), synaptophysin (1:500, Abcam, ab32127). After overnight incubation (4 °C), slides were washed with PBS and PBS-containing 0.05% Tween 20, and then incubated with appropriate secondary antibodies (Vectastatin Elite ABC Kit (Vector Laboratories, Burlingame, CA, USA) for all except pERK for which rabbit on rodent horseradish peroxidase-labeled polymer RMR622H (Biocare Medical, Concord, CA, USA) was used). Positive labeling was detected by exposing the sample to DAB+Substrate system (DAKO). Counterstaining was performed with Gill no. 3 hematoxylin (Sigma-Aldrich).
To calculate the immune cell infiltration per vision field (40 × ; 10 image per case) were included. The immunohistochemical stainings for Ki-67, CDNK2A/p16INK4a, and phospho-histone H2A.X were quantified with Immunoratio analysis software (showed as %DAB/nuclear). The immunohistochemical stainings for pERK, pS62c-Myc and YAP were quantified with ImageJ analysis software (imagej.nih.gov/ij; expressed as % positive staining per visual field with the same threshold used for each stain).
Collagen detection
Picro-Sirius Red Stain Kit (Abcam, ab150681) was used to detect collagen deposits in paraffin-embedded pancreatic tissue according to the manufacturer’s recommendations.
Real-time PCR
Total RNA was isolated using TRIzol reagent (15596-018, Invitrogen, Carlsbad, CA, USA). One microgram of total RNA was reverse transcribed using QuantiTect Reverse Transcription kit (205313, Qiagen, Valencia, CA, USA). Real-time quantitative PCR using iQTM SYBR Green Supermix (170-8882, Bio-Rad, Hercules, CA, USA) or were run on the Bio-Rad iCycler. The following primers were used: IL-6 (GenBank accession 16193, Forward: 5′-GTTGCCTTCTTGGGACTGATG-3′; Reverse: 5′-ATTGCCATTGCACAACTCTTT-3′); Transforming growth factor-β (GenBank accession 21803, Forward: 5′-GAACCAAGGAGACGGAATACA G-3′; Reverse: 5′-AACCCAGGTCCTTCCTAAAGTC-3′); Fibronectin (Genbank accession 14268, Forward: 5′-TGTGGTCTACTCTGTGGGAAT G-3′; Reverse: 5′-TTGAATTGCCACCATAAGTCTG-3′); IL-1β (Genebank accession 16176, Forward: 5′-AGAGCATCCAGCTTCAAA TCT C-3′; Reverse: 5′-CAGTTG TCTAATGGGAACGTCA-3′), RPS6 (GenBank accession 20104, Forward: 5′-GATGATGTCCGCCAGTATGTT-3′; Reverse: 5′-TTGTTCTTCTTAGTGCGTTGC-3′).
Detection of K-ras mutations by qPCR
The tumor cells from PDAC areas of PC were microdissected from paraffin-embedded tissue sections and placed in tubes and xylene was added and centrifuged. Genomic DNA was extracted from the microdissected tumor cells using standard QIAamp DNA FFPE Tissue Kit (Qiagen, 56404) according to the manufacturer’s recommendations. The qPCR was performed to detect K-ras mutation using the LNA-based probe and conditions previously described.10 The sequence of primers used: K-ras (F: aggcctgctgaaaatgactg, R: tctatcgtagggtcgtactcatc, wtLNA: cctacgccaccagctcc-PH; TIB Molbiol, LLC, Adelphia, NJ, USA). For PCR, 20ng of genomic DNA, 0.5 μm of each primers and 0.25 μm of LNA, and SensiMIX Sybr No-Rox Kit (Bioline, Taunton, MA, USA) and water were added to the final volume of 20 μl/per reaction. PCR was performed on LightCycler480 (Roche, Indianapolis, IN, USA). gDNA samples that contain K-ras mutations (extracted from tumors of mice carrying K-ras G12D and G12V mutations) and gDNA of wild-type mice (without K-ras mutation) were used as a positive and negative controls, respectively.
Sequencing of PCR products
We performed direct DNA sequencing of all tumor samples. In brief, genomic DNA was isolated from microdissected tumor samples of different PC subtypes, then, a primer pair of forward sequence: gataaagtttttgataatcttgtgtgagac and reverse sequence: atagatcaaaggcaaagctatattcttaac was used to attempt to amplify K-ras fragments. The PCR products were purified using Qiagen PCR purification kit, and TA ligated into pCR4-TOPO vector (SKU K4595-01, Thermofisher Scientific, Waltham, MA, USA) and then transfected into Escherichia coli. For each samples, we sequenced >20 clones using M13F primer.
Western blot analysis
Pancreas tissues were homogenized in lysis buffer and immunoblot analysis was performed for primary antibodies: pIKKα/β (1:1000, CST, 2697P), COX-2 (1: 2000, CST, 12282S) and GAPDH (1:10 000; Sigma-Aldrich). Briefly, proteins were separated using SDS–PAGE (polyacrylamide gel) gels, transferred to nitrocellulose membranes, and blocked with 5% non-fat milk in PBS-containing 0.05% Tween 20 for 1 h. After overnight incubation (4 °C) with primary antibodies, appropriate secondary antibody was used, and the visualization of protein bands was performed with Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NA, USA).
Statistical analysis
Statistical analysis was performed using the Prism 5 software program (GraphPad Software, San Diego, CA, USA). The percentage of survival was analyzed using the Mantel–Cox method. The median survival in days was presented. The students t-test (two-tailed, two sample equal variance) was used to compare variables between groups. The mean and s.d. values are presented. The P-value <0.05 was considered significant.
References
Balkwill F, Mantovani A . Inflammation and cancer: back to Virchow? Lancet 2001; 357: 539–545.
Ryan DP, Hong TS, Bardessy N . Pancreatic adenocarcinoma. N Engl J Med 2014; 371: 1039–1049.
Cho CH, Yu J . From inflammation to cancer. Advances in Diagnosis and Therapy for Gastrointestinal and Hepatological Diseases. World Scientific Publishing Co.: Singapore, 2012.
Murata M, Thanan R, Ma N, Kawanishi S . Role of nitrative and oxidative DNA damage in inflammation-related carcinogenesis. J Biomed Biotechnol 2012; 2012: 623019.
Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A . Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 2009; 7: 1073–1081.
Ji B, Song J, Tsou L, Bi Y, Gaiser S, Mortensen R, Logsdon C . Robust acinar cell transgene expression of CreErT via BAC recombineering. Genesis 2008; 46: 390–395.
Wood LD, Klimstra DS . Pathology and genetics of pancreatic neoplasms with acinar differentiation. Semin Diagn Pathol 2014; 31: 491–497.
La Rosa S, Sessa F, Capella C . Acinar cell carcinoma of the pancreas: overview of clinicopathologic features and insights into the molecular pathology. Front Med 2015; 2: 41.
Holmes C, Stanford WL . Concise review: stem cell antigen-1: expression, function, and enigma. Stem Cells 2007; 25: 1339–1347.
Morton JP, Klimstra DS, Mongeau ME, Lewis BC . TrTP53 deletion stimulates the formation of metastatic pancreatic tumors. Am J Pathol 2008; 172: 1081–1087.
Hu Y, Le Leu RK, Young GP . Detection of K-ras mutations in azoxymethane-induced aberrant crypt foci in mice using LNA-mediated real-time PCR clamping and mutant-specific probes. Mutat Res 2009; 677: 27–32.
Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF et al. Both p16(Ink4a) and the p19(Arf)-TP53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA 2006; 103: 5947–5952.
Morvaridi S, Dhall D, Greene MI, Pandol SJ, Wang Q . Role of YAP and TAZ in pancreatic ductal adenocarcinoma and in stellate cells associated with cancer and chronic pancreatitis. Sci Rep 2015; 5: 16759.
Phesse TJ, Myant KB, Cole AM, Ridgway RA, Ridgway RA, Pearson H et al. Endogenous c-Myc is essential for TP53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ 2014; 21: 956–966.
Grippo PJ, Sandgren EP . Acinar-to-ductal metaplasia accompanies c-myc-induced exocrine pancreatic cancer progression in transgenic rodents. Int J Cancer 2012; 131: 1242–1248.
Myant K, Qiao X, Halonen T, Come C, Laine A, Janghorban M et al. Serine 62-phosphorylated MYC associates with nuclear lamins and its regulation by CIP2A is essential for regenerative proliferation. Cell Rep 2015; 12: 1019–1031.
Hill R, Calvopina JH, Kim C, Wang Y, Dawson DW, Donahue TR et al. PTEN loss accelerates KrasG12D-induced pancreatic cancer development. Cancer Res 2010; 70: 7114–7124.
Huang H, Liu Y, Daniluk J, Gaiser S, Chu J, Wang H et al. Activation of nuclear factor-κB in acinar cells increases the severity of pancreatitis in mice. Gastroenterology 2013; 144: 202–210.
Wang W, Abbruzzese JL, Evans DB, Larry L, Cleary KR, Chiao PJ . The nuclear factor-kappa B RelA transcription factor is constitutively activated in human pancreatic adenocarcinoma cells. Clin Cancer Res 1999; 5: 119–127.
Ling S, Feng T, Jia K, Tian Y, Li Y . Inflammation to cancer: the molecular biology in the pancreas (Review). Oncol Lett 2014; 7: 1747–1754.
Liu B, Qu L, Yan S . Cyclooxygenase-2 promotes tumor growth and suppresses tumor immunity. Cancer Cell Int 2015; 15: 106.
Ricciotti E, FitzGerald GA . Prostaglandins and inflammation. Arterioscler Thromb Vasc 2011; 31: 986–1000.
Colby JK, Klein RD, McArthur MJ, Conti CJ, Kiguchi K, Kawamoto T et al. Progressive metaplastic and dysplastic changes in mouse pancreas induced by cyclooxygenase-2 overexpression. Neoplasia 2008; 10: 782–796.
Sandgren EP, Luetteke NC, Palmiter RD, Brinster RL, Lee DC . Overexpression of TGF alpha in transgenic mice: induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. Cell 1990; 61: 1121–1135.
Marrache F, Tu SP, Bhagat G, Pendyala S, Osterreicher CH, Gordon S et al. Overexpression of interleukin-1beta in the murine pancreas results in chronic pancreatitis. Gastroenterology 2008; 135: 1277–1287.
Urban T, Ricci S, Grange JD, Lacave R, Boudghene F, Breittmayer F et al. Detection of c-Ki-ras mutation by PCR/RFLP analysis and diagnosis of pancreatic adenocarcinomas. J Natl Cancer Inst 1993; 85: 2008–2012.
Kwon MJ, Jeon JY, Park HR, Nam ES, Cho SJ, Shin HS et al. Low frequency of KRAS mutation in pancreatic ductal adenocarcinomas in Korean patients and its prognostic value. Pancreas 2015; 44: 484–492.
Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ . Drugging the undruggable Ras: mission possible? Nat Rev Drug Disc 2014; 13: 828–851.
Feldmann G, Karikari C, dal Molin M, Duringer S, Volkmann P, Bartsch DK et al. Inactivation of Brca2 cooperates with TrTP53(R172H) to induce invasive pancreatic ductal adenocarcinomas in mice: a mouse model of familial pancreatic cancer. Cancer Biol Ther 2011; 11: 959–968.
Rowley M, Ohashi A, Mondal G, Mills L, Yang L, Zhang L et al. Inactivation of Brca2 promotes TrTP53-associated but inhibits KrasG12D-dependent pancreatic cancer development in mice. Gastroenterology 2011; 140: 1303–1313.
Jiao Y, Yonescu R, Offerhaus GJ, Klimstra DS, Maitra A, Eshleman JR et al. Whole-exome sequencing of pancreatic neoplasms with acinar differentiation. J Pathol 2014; 232: 428–435.
La Rosa S, Bernasconi B, Frattini M, Tibiletti MG, Molinari F, Furlan D et al. TP53 alterations in pancreatic acinar cell carcinoma: new insights into the molecular pathology of this rare cancer. Virchows Arch 2015; 468: 289–296.
Guerra C, Schuhmacher AJ, Cañamero M, Grippo PJ, Verdaguer L, Pérez-Gallego L et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 2007; 11: 291–302.
Huang H, Daniluk J, Liu Y, Chu J, Li Z, Ji B et al. Oncogenic K-Ras requires activation for enhanced activity. Oncogene 2014; 33: 532–535.
Daniluk J, Liu Y, Deng D, Chu J, Huang H, Gaiser S et al. An NF-ĸB pathway-mediated positive feedback loop amplifies Ras activity to pathological levels in mice. J Clin Invest 2012; 122: 1519–1528.
Philip B, Roland CL, Daniluk J, Liu Y, Chatterjee D, Gomez SB et al. A high-fat diet activates oncogenic Kras and COX2 to induce development of pancreatic ductal adenocarcinoma in mice. Gastroenterology 2013; 145: 1449–1458.
Kamei K, Ishikawa TO, Herschman HR . Transgenic mouse for conditional, tissue-specific Cox-2 overexpression. Genesis 2006; 44: 177–182.
Jonkers J, Meuwissen R, van der GH, Peterse H, van der Valk M, Berns A . Synergistic tumor suppressor activity of BRCA2 and TP53 in a conditional mouse model for breast cancer. Nat Genet 2001; 29: 418–425.
Acknowledgements
We thank Dr Bogdan Czerniak, Dr Tadeusz Majewski and Jolanta Bondaruk from Department of Pathology and Todd Moore from Department of Surgery, University of Texas, M.D. Anderson Cancer Center, Houston, TX, USA for technical support. Grant support: NIH-DK05206, AA020822 and The Lockton Endowment (to CDL).
Author contributions
Study concept and design: ASS, ZCM, BJ and CDL. Study supervision: CDL. Acquisition of data: ASS, ZCM, SGG, DD, HH, BJ and CDL. Analysis and interpreting the results: ASS, SGG, ZCM, LY, DD, HH, BJ, JD, WL, NA and CDL. Technical and material support: LY, JD, NA and WL. Histological characterization and analysis of the pancreatic lesions: HW and AM. Drafting of the manuscript, critical revision for important intellectual content: ASS and CDL. Obtainment of funding: CDL. All authors revised and approved the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Swidnicka-Siergiejko, A., Gomez-Chou, S., Cruz-Monserrate, Z. et al. Chronic inflammation initiates multiple forms of K-Ras-independent mouse pancreatic cancer in the absence of TP53. Oncogene 36, 3149–3158 (2017). https://doi.org/10.1038/onc.2016.461
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.461
This article is cited by
-
Molecular and metabolic regulation of immunosuppression in metastatic pancreatic ductal adenocarcinoma
Molecular Cancer (2023)
-
Cancer-inducing niche: the force of chronic inflammation
British Journal of Cancer (2022)
-
Connecting the Human Microbiome and Pancreatic Cancer
Cancer and Metastasis Reviews (2022)
-
Epigenetic inactivation of the autophagy–lysosomal system in appendix in Parkinson’s disease
Nature Communications (2021)
-
Beyond just a tight fortress: contribution of stroma to epithelial-mesenchymal transition in pancreatic cancer
Signal Transduction and Targeted Therapy (2020)