Abstract
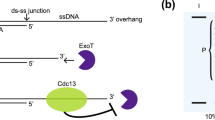
Telomere capping conceals chromosome ends from exonucleases and checkpoints, but the full range of capping mechanisms is not well defined. Telomeres have the potential to form G-quadruplex (G4) DNA, although evidence for telomere G4 DNA function in vivo is limited. In budding yeast, capping requires the Cdc13 protein and is lost at nonpermissive temperatures in cdc13-1 mutants. Here, we use several independent G4 DNA–stabilizing treatments to suppress cdc13-1 capping defects. These include overexpression of three different G4 DNA binding proteins, loss of the G4 DNA unwinding helicase Sgs1, or treatment with small molecule G4 DNA ligands. In vitro, we show that protein-bound G4 DNA at a 3′ overhang inhibits 5′→3′ resection of a paired strand by exonuclease I. These findings demonstrate that, at least in the absence of full natural capping, G4 DNA can play a positive role at telomeres in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Palm, W. & de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 (2008).
Linger, B.R. & Price, C.M. Conservation of telomere protein complexes: shuffling through evolution. Crit. Rev. Biochem. Mol. Biol. 44, 434–446 (2009).
Garvik, B., Carson, M. & Hartwell, L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15, 6128–6138 (1995).
Grandin, N., Damon, C. & Charbonneau, M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20, 1173–1183 (2001).
Nugent, C.I., Hughes, T.R., Lue, N.F. & Lundblad, V. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274, 249–252 (1996).
Burge, S., Parkinson, G.N., Hazel, P., Todd, A.K. & Neidle, S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 34, 5402–5415 (2006).
Risitano, A. & Fox, K.R. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 32, 2598–2606 (2004).
Risitano, A. & Fox, K.R. Stability of intramolecular DNA quadruplexes: comparison with DNA duplexes. Biochemistry 42, 6507–6513 (2003).
Venczel, E.A. & Sen, D. Parallel and antiparallel G-DNA structures from a complex telomeric sequence. Biochemistry 32, 6220–6228 (1993).
Paeschke, K., Simonson, T., Postberg, J., Rhodes, D. & Lipps, H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 12, 847–854 (2005).
Zhang, M.L. et al. Yeast telomerase subunit Est1p has guanine quadruplex-promoting activity that is required for telomere elongation. Nat. Struct. Mol. Biol. 17, 202–209 (2010).
Zubko, M.K., Guillard, S. & Lydall, D. Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13–1 mutants. Genetics 168, 103–115 (2004).
Hayashi, N. & Murakami, S. STM1, a gene which encodes a guanine quadruplex binding protein, interacts with CDC13 in Saccharomyces cerevisiae. Mol. Genet. Genomics 267, 806–813 (2002).
Frantz, J.D. & Gilbert, W. A yeast gene product, G4p2, with a specific affinity for quadruplex nucleic acids. J. Biol. Chem. 270, 9413–9419 (1995).
Van Dyke, M.W., Nelson, L.D., Weilbaecher, R.G. & Mehta, D.V. Stm1p, a G4 quadruplex and purine motif triplex nucleic acid-binding protein, interacts with ribosomes and subtelomeric Y′ DNA in Saccharomyces cerevisiae. J. Biol. Chem. 279, 24323–24333 (2004).
Nelson, L.D., Musso, M. & Van Dyke, M.W. The yeast STM1 gene encodes a purine motif triple helical DNA-binding protein. J. Biol. Chem. 275, 5573–5581 (2000).
Zubko, M.K. & Lydall, D. Linear chromosome maintenance in the absence of essential telomere-capping proteins. Nat. Cell Biol. 8, 734–740 (2006).
Huber, M.D., Lee, D.C. & Maizels, N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 30, 3954–3961 (2002).
Huber, M.D., Duquette, M.L., Shiels, J.C. & Maizels, N.A. Conserved G4 DNA binding domain in RecQ family helicases. J. Mol. Biol. 358, 1071–1080 (2006).
Vodenicharov, M.D. & Wellinger, R.J. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol. Cell 24, 127–137 (2006).
Fernando, H. et al. Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 37, 6716–6722 (2009).
Piazza, A. et al. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 38, 4337–4348 (2010).
De Cian, A., DeLemos, E., Mergny, J.-L., Teulade-Fichou, M.-P. & Monchaud, D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 129, 1856–1857 (2007).
Ren, J. & Chaires, J.B. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry 38, 16067–16075 (1999).
Smith, J.S. & Johnson, F.B. Isolation of G-quadruplex DNA using NMM-sepharose affinity chromatography. Methods Mol. Biol. 608, 207–221 (2010).
Li, Y., Geyer, R. & Sen, D. Recognition of anionic porphyrins by DNA aptamers. Biochemistry 35, 6911–6922 (1996).
Hershman, S.G. et al. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 36, 144–156 (2008).
Zhu, Z., Chung, W.-H., Shim, E.Y., Lee, S.E. & Ira, G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 (2008).
Watt, P.M., Hickson, I.D., Borts, R.H. & Louis, E.J. SGS1, a homologue of the Bloom's and Werner's Syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144, 935–945 (1996).
Mimitou, E.P. & Symington, L.S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 (2008).
Frei, C. & Gasser, S. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 14, 81–96 (2000).
Cobb, J.A., Bjergbaek, L., Shimada, K., Frei, C. & Gasser, S.M. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22, 4325–4336 (2003).
Bernstein, D.A. & Keck, J.L. Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure 13, 1173–1182 (2005).
Mullen, J.R., Kaliraman, V. & Brill, S.J. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154, 1101–1114 (2000).
Liu, Z. et al. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure 7, 1557–1566 (1999).
Lu, J. et al. Human homologues of yeast helicase. Nature 383, 678–679 (1996).
Maringele, L. & Lydall, D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 16, 1919–1933 (2002).
Lin, J., Smith, D.L. & Blackburn, E.H. Mutant telomere sequences lead to impaired chromosome separation and a unique checkpoint response. Mol. Biol. Cell 15, 1623–1634 (2004).
Lane, A.N., Chaires, J.B., Gray, R.D. & Trent, J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 36, 5482–5515 (2008).
Förstemann, K., Hoss, M. & Lingner, J. Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res. 28, 2690–2694 (2000).
Saccà, B., Lacroix, L. & Mergny, J.-L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 33, 1182–1192 (2005).
Mergny, J.-L., Li, J., Lacroix, L., Amrane, S. & Chaires, J.B. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 33, e138 (2005).
Kypr, J., Kejnovska, I., Renciuk, D. & Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 37, 1713–1725 (2009).
Sandell, L.L. & Zakian, V.A. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75, 729–739 (1993).
Addinall, S.G. et al. A genomewide suppressor and enhancer analysis of cdc13–1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics 180, 2251–2266 (2008).
Downey, M. et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124, 1155–1168 (2006).
Ribeyre, C. et al. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 5, e1000475 (2009).
Vallur, A.C. & Maizels, N. Distinct activities of exonuclease 1 and flap endonuclease 1 at telomeric G4 DNA. PLoS ONE 5, e8908 (2010).
Salas, T.R. et al. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 34, 4857–4865 (2006).
Tsai, Y.-C., Qi, H. & Liu, L.F. Protection of DNA ends by telomeric 3′ G-tail sequences. J. Biol. Chem. 282, 18786–18792 (2007).
Michelson, R.J., Rosenstein, S. & Weinert, T. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 19, 2546–2559 (2005).
Bonetti, D., Martina, M., Clerici, M., Lucchini, G. & Longhese, M.P. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol. Cell 35, 70–81 (2009).
Giraldo, R. & Rhodes, D. The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J. 13, 2411–2420 (1994).
Pedroso, I.M., Hayward, W. & Fletcher, T.M. The effect of the TRF2 N-terminal and TRFH regions on telomeric G-quadruplex structures. Nucleic Acids Res. 37, 1541–1554 (2009).
Sfeir, A. et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138, 90–103 (2009).
Ding, H. et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell 117, 873–886 (2004).
Gomez, D. et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 66, 6908–6912 (2006).
Phatak, P. et al. Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. Br. J. Cancer 96, 1223–1233 (2007).
Oganesian, L., Graham, M.E., Robinson, P.J. & Bryan, T.M. Telomerase recognizes G-quadruplex and linear DNA as distinct substrates. Biochemistry 46, 11279–11290 (2007).
Kozak, M.L. et al. Inactivation of the Sas2 histone acetyltransferase delays senescence driven by telomere dysfunction. EMBO J. 29, 158–170 (2010).
Acknowledgements
We thank the members of the Johnson and Yatsunyk labs for helpful discussions and comments on the manuscript; N. Maizels, M. Fry, K. Runge, V. Zakian, J.-L. Mergny, S. Berger, R. Marmorstein and P. Adams for discussions; S. Murakami (Kanazawa University), E. Blackburn (University of California, San Francisco), M. Charbonneau (École Normale Supérieure), S. Brill (Rutgers University), S. Gasser (Friedrich Miescher Institute) and M. Van Dyke (MD Anderson Cancer Center) for providing strains and plasmids, and D. Durocher (University of Toronto) for the Rad53 antibody. This work was supported by US National Institutes of Health grants R01 AG021521 (F.B.J.), P01 AG031862 (F.B.J.), T32 GM008216-22 (J.S.S.) and T32 GM07229 (J.S.S.), a Camille and Henry Dreyfus Faculty Startup Award (L.A.Y.), Research Corporation Award no. 7843 (L.A.Y.), a Deutscher Akademischer Austausch Dienst Fellowship (R.K.) and by a Cancer Research UK program grant (S.B.).
Author information
Authors and Affiliations
Contributions
J.S.S. and F.B.J. conceived of and carried out experiments, interpreted results and wrote the manuscript. L.A.Y. and J.M.N. conducted and designed CD, thermal difference spectroscopy and fluorescence resonance energy transfer experiments, along with F.B.J. and J.S.S., and provided comments on the manuscript. R.K. and S.B. provided the HF1 cDNA and protein, designed and conducted CD and ELISA experiments (Supplementary Fig. 3) and provided comments on the manuscript. D.C.S. provided purified proteins and comments on the manuscript. D.M. and M.-P. T.-F. synthesized the bisquinolinium G4 DNA ligands and provided comments on the manuscript. Q.C., M.S.G. and L.A. carried out experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Table 1 and Supplementary Methods (PDF 4612 kb)
Rights and permissions
About this article
Cite this article
Smith, J., Chen, Q., Yatsunyk, L. et al. Rudimentary G-quadruplex–based telomere capping in Saccharomyces cerevisiae. Nat Struct Mol Biol 18, 478–485 (2011). https://doi.org/10.1038/nsmb.2033
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2033
This article is cited by
-
Dynamic interaction of BRCA2 with telomeric G-quadruplexes underlies telomere replication homeostasis
Nature Communications (2022)
-
Telomerase subunit Est2 marks internal sites that are prone to accumulate DNA damage
BMC Biology (2021)
-
Characterization of metapopulation of Ellobium chinense through Pleistocene expansions and four covariate COI guanine-hotspots linked to G-quadruplex conformation
Scientific Reports (2021)
-
Extraordinary diversity of telomeres, telomerase RNAs and their template regions in Saccharomycetaceae
Scientific Reports (2021)
-
The regulation and functions of DNA and RNA G-quadruplexes
Nature Reviews Molecular Cell Biology (2020)