Abstract
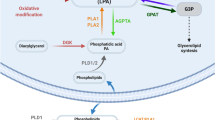
Autotaxin (ATX, also known as ectonucleotide pyrophosphatase/phosphodiesterase-2, ENPP2) is a secreted lysophospholipase D that generates the lipid mediator lysophosphatidic acid (LPA), a mitogen and chemoattractant for many cell types. ATX-LPA signaling is involved in various pathologies including tumor progression and inflammation. However, the molecular basis of substrate recognition and catalysis by ATX and the mechanism by which it interacts with target cells are unclear. Here, we present the crystal structure of ATX, alone and in complex with a small-molecule inhibitor. We have identified a hydrophobic lipid-binding pocket and mapped key residues for catalysis and selection between nucleotide and phospholipid substrates. We have shown that ATX interacts with cell-surface integrins through its N-terminal somatomedin B–like domains, using an atypical mechanism. Our results define determinants of substrate discrimination by the ENPP family, suggest how ATX promotes localized LPA signaling and suggest new approaches for targeting ATX with small-molecule therapeutic agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Stracke, M.L. et al. Identification, purification, and partial sequence analysis of autotaxin, a novel motility-stimulating protein. J. Biol. Chem. 267, 2524–2529 (1992).
Tokumura, A. et al. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J. Biol. Chem. 277, 39436–39442 (2002).
Umezu-Goto, M. et al. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J. Cell Biol. 158, 227–233 (2002).
Noguchi, K., Herr, D., Mutoh, T. & Chun, J. Lysophosphatidic acid (LPA) and its receptors. Curr. Opin. Pharmacol. 9, 15–23 (2009).
van Meeteren, L.A. & Moolenaar, W.H. Regulation and biological activities of the autotaxin-LPA axis. Prog. Lipid Res. 46, 145–160 (2007).
van Meeteren, L.A. et al. Autotaxin, a secreted lysophospholipase D, is essential for blood vessel formation during development. Mol. Cell. Biol. 26, 5015–5022 (2006).
Tanaka, M. et al. Autotaxin stabilizes blood vessels and is required for embryonic vasculature by producing lysophosphatidic acid. J. Biol. Chem. 281, 25822–25830 (2006).
Koike, S. et al. Autotaxin/lysophospholipase D-mediated lysophosphatidic acid signaling is required to form distinctive large lysosomes in the visceral endoderm cells of the mouse yolk sac. J. Biol. Chem. 284, 33561–33570 (2009).
Fotopoulou, S. et al. ATX expression and LPA signalling are vital for the development of the nervous system. Dev. Biol. 339, 451–464 (2010).
Lin, S. et al. The absence of LPA2 attenuates tumor formation in an experimental model of colitis-associated cancer. Gastroenterology 136, 1711–1720 (2009).
Liu, S. et al. Expression of autotaxin and lysophosphatidic acid receptors increases mammary tumorigenesis, invasion, and metastases. Cancer Cell 15, 539–550 (2009).
Mills, G.B. & Moolenaar, W.H. The emerging role of lysophosphatidic acid in cancer. Nat. Rev. Cancer 3, 582–591 (2003).
Nam, S.W. et al. Autotaxin (ATX), a potent tumor motogen, augments invasive and metastatic potential of ras-transformed cells. Oncogene 19, 241–247 (2000).
Taghavi, P. et al. In vitro genetic screen identifies a cooperative role for LPA signaling and c-Myc in cell transformation. Oncogene 27, 6806–6816 (2008).
David, M. et al. Cancer cell expression of autotaxin controls bone metastasis formation in mouse through lysophosphatidic acid-dependent activation of osteoclasts. PLoS ONE 5, e9741 (2010).
Kanda, H. et al. Autotaxin, an ectoenzyme that produces lysophosphatidic acid, promotes the entry of lymphocytes into secondary lymphoid organs. Nat. Immunol. 9, 415–423 (2008).
Pradère, J.P. et al. LPA1 receptor activation promotes renal interstitial fibrosis. J. Am. Soc. Nephrol. 18, 3110–3118 (2007).
Tager, A.M. et al. The lysophosphatidic acid receptor LPA1 links pulmonary fibrosis to lung injury by mediating fibroblast recruitment and vascular leak. Nat. Med. 14, 45–54 (2008).
Inoue, M. et al. Initiation of neuropathic pain requires lysophosphatidic acid receptor signaling. Nat. Med. 10, 712–718 (2004).
Gierse, J.K. et al. A novel autotaxin inhibitor reduces lysophosphatidic acid levels in plasma and the site of inflammation. J. Pharmacol. Exp. Ther. 334, 310–317 (2010).
Albers, H.M. et al. Boronic acid-based inhibitors of autotaxin reveals rapid turnover of LPA in the circulation. Proc. Natl. Acad. Sci. USA 107, 7257–7262 (2010).
Zhou, A. Functional structure of the somatomedin B domain of vitronectin. Protein Sci. 16, 1502–1508 (2007).
Pamuklar, Z. et al. Autotaxin/lysopholipase D and lysophosphatidic acid regulate murine hemostasis and thrombosis. J. Biol. Chem. 284, 7385–7394 (2009).
Zalatan, J.G., Fenn, T.D., Brunger, A.T. & Herschlag, D. Structural and functional comparisons of nucleotide pyrophosphatase/phosphodiesterase and alkaline phosphatase: implications for mechanism and evolution. Biochemistry 45, 9788–9803 (2006).
Jansen, S., Andries, M., Derua, R., Waelkens, E. & Bollen, M. Domain interplay mediated by an essential disulfide linkage is critical for the activity and secretion of the metastasis-promoting enzyme autotaxin. J. Biol. Chem. 284, 14296–14302 (2009).
Stefan, C., Jansen, S. & Bollen, M. NPP-type ectophosphodiesterases: unity in diversity. Trends Biochem. Sci. 30, 542–550 (2005).
Jansen, S. et al. An essential oligomannosidic glycan chain in the catalytic domain of autotaxin, a secreted lysophospholipase-D. J. Biol. Chem. 282, 11084–11091 (2007).
Day, J.E. et al. Crystallization and preliminary X-ray diffraction analysis of rat autotaxin. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1127–1129 (2010).
Hausmann, J. et al. Mammalian cell expression, purification, crystallization and microcrystal data collection of autotaxin/ENPP2, a secreted mammalian glycoprotein. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1130–1135 (2010).
Ghosh, M., Meiss, G., Pingoud, A., London, R.E. & Pedersen, L.C. Structural insights into the mechanism of nuclease A, a ββα metal nuclease from Anabaena. J. Biol. Chem. 280, 27990–27997 (2005).
Bollen, M., Gijsbers, R., Ceulemans, H., Stalmans, W. & Stefan, C. Nucleotide pyrophosphatases/phosphodiesterases on the move. Crit. Rev. Biochem. Mol. Biol. 35, 393–432 (2000).
Lee, J. et al. Enzymatic activation of autotaxin by divalent cations without EF-hand loop region involvement. Biochem. Pharmacol. 62, 219–224 (2001).
Albers, H.M. et al. Discovery and optimization of boronic acid based inhibitors of autotaxin. J. Med. Chem. 53, 4958–4967 (2010).
Golovin, A. & Henrick, K. MSDmotif: exploring protein sites and motifs. BMC Bioinformatics 9, 312 (2008).
de Vries, S.J., van Dijk, M. & Bonvin, A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5, 883–897 (2010).
Cimpean, A., Stefan, C., Gijsbers, R., Stalmans, W. & Bollen, M. Substrate-specifying determinants of the nucleotide pyrophosphatases/phosphodiesterases NPP1 and NPP2. Biochem. J. 381, 71–77 (2004).
Zhou, A., Huntington, J.A., Pannu, N.S., Carrell, R.W. & Read, R.J. How vitronectin binds PAI-1 to modulate fibrinolysis and cell migration. Nat. Struct. Biol. 10, 541–544 (2003).
Huai, Q. et al. Crystal structures of two human vitronectin, urokinase and urokinase receptor complexes. Nat. Struct. Mol. Biol. 15, 422–423 (2008).
Li, X., Zou, G., Yuan, W. & Lu, W. Defining the native disulfide topology in the somatomedin B domain of human vitronectin. J. Biol. Chem. 282, 5318–5326 (2007).
van Meeteren, L.A. et al. Inhibition of autotaxin by lysophosphatidic acid and sphingosine 1-phosphate. J. Biol. Chem. 280, 21155–21161 (2005).
Eichholtz, T., Jalink, K., Fahrenfort, I. & Moolenaar, W.H. The bioactive phospholipid lysophosphatidic acid is released from activated platelets. Biochem. J. 291, 677–680 (1993).
Prudente, S., Morini, E. & Trischitta, V. Insulin signaling regulating genes: effect on T2DM and cardiovascular risk. Nat Rev Endocrinol 5, 682–693 (2009).
McCoy, A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007).
Langer, G., Cohen, S.X., Lamzin, V.S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 (2004).
Schüttelkopf, A.W. & van Aalten, D.M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 (2004).
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Acknowledgements
Crystallographic experiments were conducted at the PX beamline at the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland; at the European Synchrotron Radiation Facility beamline ID23-2; at the Diamond Light Source ID19 microfocus beamline; and at the GM/CA-CAT, NE-CAT and LS-CAT beamlines at the Advanced Photon Source. We thank all beamline scientists and especially S. McSweeney, D. Flo, K. Schultze-Briese, T. Tomizaki, G. Evans and J. Grimes for data collection assistance. This research was supported by grants from the Dutch Cancer Society to W.H.M., the US National Institutes of Health to A.J.M. (GM50388, P20RR021954), C.V.K. (GM094155) and S.S.S. (HL078663), F30 HL099272 to Z.F. and an American Heart Association postdoctoral fellowship to T.W. Material for these authors is the result of work supported with the resources and use of the facilities at the Lexington Veterans Affairs Medical Center. E.C. and A.P. thank V. De Marco, D. Littler and P. Rucktooa for assistance with crystal mounting and data collection. J.H. and A.P. thank T. Walter, P. Celie, M. Sturnaiolo and T. Heidebrecht for advice and assistance, the Netherlands Cancer Institute and Pfizer for jointly providing a pre-doctoral fellowship to J.H., T. Sixma for feedback on the manuscript and the NKI Protein Facility for infrastructure access.
Author information
Authors and Affiliations
Contributions
J.H. purified, crystallized and collected diffraction data for native crystal forms; established co-crystals, collected diffraction data and participated in structure determination of the HA155 complex; and cloned, expressed and assayed activity of catalytic and binding site mutants. S.K. directly supervised Pfizer scientists, collected and processed diffraction data, determined the first structure, built and refined crystal structures; E.C. established cell lines, protein expression and purification, crystallization protocols for the first native crystal form, and collected diffraction data; J.E.D. identified and produced the second, high resolution, native crystal form and collected diffraction data; T.W. and Z.F. generated all reagents for ATX platelet binding experiments and did the relevant assays; L.A.v.M. participated in protein expression and with the help of H.M.H.G.A. and A.J.S.H., established ATX assays; H.M.H.G.A. made the HA155 inhibitor; L.v.Z. conducted the PAI-1 experiment; S.J. generated the rat ATX mutant used for crystallization, M.A. did the EF hand–like mutagenesis experiments and assays; T.H. purified protein; L.E.P. expressed protein; T.E.B. coordinated the Pfizer scientists; K.H. assisted with crystal growth, manipulation and data collection; M.K. helped establish the stable cell line producing rat ATX; C.W.V.K., S.S.S. and A.J.M. designed and supervised platelet binding experiments; H.O. supervised inhibitor design at the NKI site; M.B. supervised EF-hand experiments and participated in project coordination; W.H.M. and A.P. initiated and coordinated the project and supervised experiments at the NKI. A.P. also determined and refined crystal structures, prepared all display items and wrote the paper, with input from W.H.M., S.K., M.B. and A.J.M.
Corresponding authors
Ethics declarations
Competing interests
At the time this research was performed, S.K., J.E.D., T.H., L.E.P. and T.E.B. were employees of Pfizer, Inc.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 8662 kb)
Rights and permissions
About this article
Cite this article
Hausmann, J., Kamtekar, S., Christodoulou, E. et al. Structural basis of substrate discrimination and integrin binding by autotaxin. Nat Struct Mol Biol 18, 198–204 (2011). https://doi.org/10.1038/nsmb.1980
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1980
This article is cited by
-
Autotaxin inhibitor IOA-289 reduces gastrointestinal cancer progression in preclinical models
Journal of Experimental & Clinical Cancer Research (2023)
-
Lipid phosphate phosphatase 3 in smooth muscle cells regulates angiotensin II-induced abdominal aortic aneurysm formation
Scientific Reports (2022)
-
AgRP neurons control feeding behaviour at cortical synapses via peripherally derived lysophospholipids
Nature Metabolism (2022)
-
Prevention of age-associated neuronal hyperexcitability with improved learning and attention upon knockout or antagonism of LPAR2
Cellular and Molecular Life Sciences (2021)
-
Pleotropic Roles of Autotaxin in the Nervous System Present Opportunities for the Development of Novel Therapeutics for Neurological Diseases
Molecular Neurobiology (2020)